An Isozyme Marker Linked to the N-1 Gene Governing Nakedness in Oat
Solomon
Kibite
Agriculture and Agri-Food Canada, Lacombe Research Centre, 6000 C & E Trail, Lacombe, Alberta T0C 1S0, Canada
Two types of oat, covered oats and naked oats, are produced and marketed in western Canada. Covered oats account for more than 95% of the oat production in Alberta, Saskatchewan and Manitoba, but interest in naked oats is gradually increasing as farmers continue to search for alternative sources of nutritious and inexpensive feed for their livestock. This has stimulated interest among oat breeders to develop high yielding, disease resistant naked oat cultivars for western Canada.
Kernel nakedness is a genetically complex trait in oats; alleles at four different loci (designated as N-1, N-2, N-3 and N-4?) are known to act epistatically to govern its expression (2). N-1 is the major switch gene conditioning nakedness (1), and alleles of the other three loci interact with N-1 and among each other to modify the degree to which nakedness is expressed. Depending on the alleles present at each the four loci, oat genotypes demonstrate different degrees of nakedness ranging from completely naked (or nearly naked) to partially naked, to partially covered, and all the way to completely covered phenotypes. A completely naked phenotype is expressed when dominant alleles are present at the N-1 and N-2 loci, and either the dominant (N-3) or the recessive (n-3) allele is present at the third locus. Genotypes with homozygous recessive (n-1/n-1) alleles at the N-1 locus will always have the covered phenotype regards of the alleles present at the other loci. N-1/N-1 genotypes may have either naked or mosaic phenotypes depending on the alleles present at the other loci. The mosaic phenotype produces a mixture of naked and covered kernels, of various proportions, depending on the alleles present at the N-2 and N-3 loci. The N-4? locus reverses the effects of the other three loci. In a homozygous recessive (n-4/n-4) condition it is hypostatic to any dominant allele at the other three loci. When the N-4 locus is homozygous dominant (N-4/N-4) and the N-1 locus is in a heterozygous condition, a covered phenotype is produced.
Hull retention, which is normally related to the expression of the mosaic phenotype, is a major grain quality problem in naked oats. Since hull retention affects the market value and the nutritional quality of the grain, most oat breeders are interested in developing free-threshing naked oat cultivars with minimal hull retention characteristics. Breeding for free-threshing naked oat cultivars would be simplified if methods were developed that would enable plant breeders to select for the best combinations of alleles at the N-1, N-2, N-3 and N-4 loci. We began this study to develop markers that can be used as tools to select against the complex inter-allelic interactions that govern the expression of hull retention in naked oats. This paper describes the development of an esterase isozyme marker that is tightly linked to one of the genes (presumably N-1) conditioning nakedness in oats.
The isozyme marker was discovered using 8 pairs of covered and naked near-isogenic lines. The 8 pairs of near-isogenic lines were developed from a cross of a naked oat genotype, OT253, and a covered oat cultivar, Marion. The following narrative describes the development of the near-isogenic lines and the discovery of the marker.
The F1 generation of the OT253/Marion cross was grown in a growth chamber. The F2 through F6 generations were developed in a greenhouse by single-seed descent. A single panicle was harvested from each F6 plant and was grown in a panicle-to-row nursery to produce F6:7 families. Each of the F6:7 families consisted of approximately 20 - 30 individuals. At heading, the families were classified as homozygous covered, homozygous naked or segregating based on panicle and kernel characteristics. From an initial stock of approximately 200 F6:7 families, eight families segregating for kernel characteristics (i.e. covered vs. naked) were identified. From each of these eight families 10 panicles harvested at random. These random panicles were threshed individually and planted in pots (20 kernels/family) in a greenhouse in an effort to advance the generations of the 8 families by maintaining heterozygosity. This procedure was repeated for two more generations until the F6:10 generation was attained. The F6:10 generation was grown in a panicle-to-row nursery. The nursery was established on a Ponoka clay-loam soil at the Lacombe Research Centre during the 1995 growing season. Throughout the growing season the F6:10 families were scored for uniformity, panicle characteristics and kernel type. From each of the eight groups of F6:10 family one homozygous naked and one homozygous covered family were selected. Seed from the selected families was bulked (by family) to produce the 8 pairs of near-isogenic lines. Each pair of near-isogenic lines traces back to a single F2 plant.
The 8 pairs of near-isogenic lines were subjected to isozyme analyses. Five gram seed samples of the 16 genotypes were ground in a Retsch mill, Model 2M 100, and 0.5 g of the ground samples were transferred to a polypropylene centrifuge tubes. Water-soluble proteins including esterases were extricated by addition of 2 ml of 2% glycine (pH 8.6) followed by thorough mixing and incubation at 4o C overnight. The tubes were centrifuged for 10 m at 15,000 x g. The clear supernatant of each sample was used for analysis. Samples were transferred to pre-cast gels (HyPure Gel FS-5480; pH 3 -10) by absorption of the extracts to 5 x 10 mm pieces of filter paper. The pieces of paper were loaded to the gels by insertion into wells about 20 mm from the anode. The proteins were separated by isoelectricfocusing (IEF). After 30 minutes focusing at 30 W, 1000 V maximum, the pieces of paper were removed and focusing continued until completion. Focused gels were stained either for total protein or for enzyme activity (esterase, peroxidase, acid phosphatase and phosphorylase). For esterases, gels were incubated in 100 ml of solution containing 0.1 g Fast Blue RR salt, 0.05g a-naphtylacetate (both dissolved in a small volume of acetone) and 20 ml of 0.25M phosphate buffer, pH 7.5, for about 30 minutes, with constant shaking at 30o C in the dark. Excess background stain was removed using 10% (v/v) acetic acid solution. All gels were soaked in 10% (v/v) glycerol solution for at least 1 hour and left to dry on the bench at room temperature prior to scoring.
Although the gels were stained for total protein and four enzymes, only the esterase analysis produced promising results for identification of the naked and covered phenotypes. Fig. 1 shows typical esterase banding patterns of the 8 pairs of near-isogenic lines. Each line was analyzed in duplicates; samples from covered lines were assigned odd numbers where as samples from naked lines were assigned even numbers. The covered and naked members of each isogenic pair were tested side by side on the gel. As can be seen from Fig. 1, the presence of a clearly resolved fast moving esterase band was associated with the expression of the naked phenotype. Absence of the band was associated with the covered phenotype.
To determine if the same esterase band can be used to distinguish naked oats and covered oats, 16 randomly selected naked genotypes and 16 randomly selected covered genotypes were examined. The results (Fig. 2) showed that the diagnostic band was consistently present in all of the naked lines and was consistently absent in all of the covered lines. These results suggested that the esterase isozyme was tightly linked to at least one of the genes (presumably N-1) conditioning nakedness in oats. Preliminary studies on the inheritance of the isozyme marker have suggested that it is inherited as a complex factor. These studies are continuing at the present time. Additional studies examining the effects of environmental factors on the expression of the marker are planned.
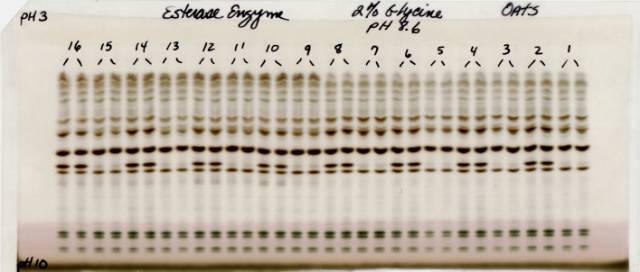
Fig. 1. Esterase banding patterns or 8 pairs of covered vs. naked near-isogenic oat lines. Odd numbered samples (i.e. 1, 3, 5, … 15) were extracted from the covered lines, and even numbered samples ( 2, 4, 6, … 16) were extracted from the naked isolines. Note that an extra band (6th dark band from the bottom) was consistently present in samples of all of the naked lines and consistently absent from samples all of the covered lines.
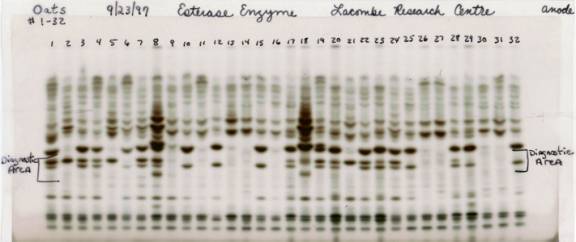
Fig. 2. Esterase banding patterns of 16 random samples of naked oats and 16 random samples of covered oats. Sample nos. 1 - 8 and 17 – 24, inclusive were naked, and samples 9 – 16 and 25 – 32, inclusive, were covered. Sample #6 (LAO-586-NZ-0268, a covered genotype) and sample # 28 (OT900, a naked genotype) were loaded in the wrong lane on the gel, but their banding patterns were consistent with those expected for a covered and a naked genotype, respectively.
References
Jenkins, G., and P.R. Hanson. 1976. The genetics of naked oats (Avena nuda L.). Euphytica 25: 167 – 174.
Simons, M.D., J.W. Martens, R.I.H. McKenzie, I. Nishiyama, K. Sadanaga, J. Sebesta and H. Thomas. 1978. Oats: A standardized system of nomenclature for genes and chromosomes and catalog of genes governing characters. USDA-SEA Agric. Handb. 509. U.S.Gov. Print. Office, Washington D.C.
Return to the Table of Contents