
Barley Genetics Newsletter 36:30-43 (2006)
New SSR markers for barley derived from the EST database
Karen A. Beaubien and Kevin P. Smith*
Department of
Agronomy and Plant Genetics,
*Corresponding author: E-mail: smith376@umn.edu
Introduction
There are currently 1,196 microsatellite [or simple sequence repeat (SSR)] marker primer sets that have been developed for barley (from both genomic libraries and EST databases) of which 504 have been mapped (Saghai Maroof et al., 1994; Becker and Heun, 1995; Liu et al., 1996; Struss and Plieske, 1998; Ramsay et al., 2000; Pillen et al., 2000; Holton et al., 2002; Thiel et al., 2003; Li et al., 2003; Yu et al., 2005). Unfortunately, the available SSR markers provide uneven coverage of the barley genome and are concentrated near the centromeres. We have compared the available mapped SSRs and identified 62 BINs out of the total of 99 barley BINs (http://barleygenomics.wsu.edu/; http://rye.pw.usda.gov/cgi-bin/gbrowse/BarleyBinMaps) that have poor coverage. Although this represents, 63% of the barley genome, only 31% of the available SSRs map to these BINs. Additional SSR markers are needed to increase coverage in these BINs. Moreover, some of the SSR markers recently published are restricted from being used to develop new barley varieties, thus there is still a need for additional publicly available SSR markers that can be used without restrictions.
Materials and Methods
Barley ESTs used for primer
development were selected by using either rice BAC or wheat EST sequences in a
BLASTn search for publicly available barley ESTs with the low complexity filter
turned off (BLASTn searches were completed between October, 2002 and October,
2004) (http://www.ncbi.nih.gov/BLAST/). Matches
with e-values between 0 and 1e-2 were used.
The resulting barley ESTs were processed through the Tandem repeats
finder which measures the rate at which the actual EST sequence matches a
perfect repeat sequence (Benson, 1999; http://tandem.bu.edu/trf/trf.submit.options.html). Primers were designed for SSRs with 85-100%
matches to the perfect repeat sequence.
Primer pairs were designed to flank SSR motifs using Primer3 software (Rozen
and Skaletsky, 2000; http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) and
screened on a set of mapping parents to identify which population(s) were
appropriate for mapping. Primer pairs
were tested using
Newly developed SSR markers were mapped on the appropriate mapping population(s) including: Steptoe x Morex (Kleinhofs et al., 1993), Chevron x M69 (Canci et al., 2003), Frederickson x Stander (Mesfin et al., 2003), or Atahualpa x M81 (unpublished). We used JoinMap 3.0 for map construction (Van Ooijen and Voorrips, 2001). Assignment to BINs was based on adjacent mapped markers that have been previously assigned to BINs (http://barleygenomics.wsu.edu/; http://rye.pw.usda.gov/cgi-bin/gbrowse/BarleyBinMaps). Wheat STS markers were mapped on the barley populations to increase the coverage of chromosome 3 (3H) (Liu and Anderson, 2003).
Results and Discussion
A total of 76 new markers were developed that produce between two to six alleles per locus among the twelve mapping parents: Atahualpa, M81, Chevron, M69, Frederickson, Stander, Harrington, OUH602, Hor211, Lacey, Steptoe and Morex (Table 3). Sixty of the markers were mapped using the Steptoe x Morex, Chevron x M69, Frederickson x Stander, and Atahualpa x M81populations (Figure 1 and Table 3). Of the 60 mapped markers, 41 (68%) have mapped to BIN positions that were previously identified as being poorly covered with the currently available SSR markers. These markers should provide additional tools for barley genetic mapping and marker-assisted selection.
Acknowledgements
The authors would like to thank Charles Gustus and Danielle Wojdyla for their assistance in STS and SSR marker data collection. This work was carried out in part using software provided by the University of Minnesota Supercomputing Center. This research was supported by U. S. Barley Genome Project.
References:
Becker, J., and M. Heun. 1995. Barley microsatellites: allele variation and mapping. Plant Mol. Biol. 27: 835-845.
Benson, G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Research 27: 573-580.
Canci,
P.C., L.M. Nduulu, R. Dill-Macky, G.J.
Holton, T.A., J.T. Christopher, L. McClure, N. Harker, and R.J. Henry. 2002. Identification and mapping of polymorphic SSr markers from expressed gene sequences of barley and wheat. Mol. Breeding 9: 63-71.
Kleinhofs, A., A. Kilian, M.A. Saghai Maroof, R.M. Biyashev, P. Hayes, F.Q. Chen, N. Lapitan, A. Fenwick, T.K. Blake, V. Kanazin, E. Ananiev, L. Dahleen, D. Kudrna, J. Bollinger, S.J. Knapp, B. Liu, M. Sorrells, M. Heun, J.D. Franckowiak, D. Hoffman, R. Skadsen, and B.J. Steffenson. 1993. A molecular, isozyme and morphological map of the barley (Hordeum vulgare) genome. Theor. Appl. Genet. 86: 705-712.
Li , J.Z., T.G. Sjakste, M.S. Röder and M.W. Ganal. 2003.
Development and genetic mapping of 127 new microsatellite markers in
barley. Theor. Appl. Genet. 107: 1021–1027.
Liu, S., and J.A. Anderson.
2003. Targeted molecular mapping
of a major wheat QTL for Fusarium head blight resistance using wheat ESTs and
synteny with rice. Genome 46: 817-823.
Liu, Z.-W., R.M. Biyashev and M.A. Saghai Maroof. 1996. Development of simple sequence repeat markers
and their integration into a barley linkage map. Theor. Appl. Genet. 93: 869–876.
Mesfin, A., K.P. Smith, R. Dill-Macky, C.K. Evans, R Waugh, C.D. Gustus, and G.J. Muehlbauer. 2003. Quantitative trait loci for Fusarium head blight resistance in barley detected in a two-rowed by six-rowed population. Crop Sci. 43: 307-318.
Pillen, K., A. Binder, B. Kreuzkam, L. Ramsay, R. Waugh, J. Förster, J. Léon. 2000. Mapping new EMBL-derived barley microsatellites and their use in differentiating German barley cultivars. Theor. Appl. Genet. 101: 652-660.
Ramsay, L., M. Macaulay, S. degli Ivanissevich, K. MacLean, L. Cardle, J. Fuller, K. J. Edwards, S. Tuvesson, M. Morgante, A. Massari, E. Maestri, N. Marmiroli, T. Sjakste, M. Ganal, W. Powell, and R. Waugh. 2000. A Simple Sequence Repeat-Based Linkage Map of Barley. Genetics 156: 1997-2005.
Rozen,
S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for
biologist programmers. In: Bioinformatics Methods and Protocols: Methods in
Molecular Biology. Eds Krawetz, S., and Misener, S. Humana.
Saghai Maroof, M.A., R.M. Biyashev, G.P. Yang, Q. Zhang and
R.W. Allard. 1994. Extraordinarily polymorphic microsatellite DNA
in barley: species diversity, chromosomal locations and population dynamics.
Proc. Natl. Acad. Sci. USA 91: 5466–5470.
Struss, D., and J. Plieske.
1998. The use of microsatellite
markers for detection of genetic diversity in barley populations. Theor.
Appl. Genet. 97: 308-15.
Thiel, T, W. Michalek, R.K. Varshney, and A. Graner. 2003. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor. Appl. Genet. 106: 411–422.
Van
Ooijen, J.W., and R.E. Voorrips.
2001. JoinMap 3.0, Software for
the calculation of genetic linkage maps.
Plant Research International, Wageningen, the
Table 1. PCR recipes for UMB SSR markers
|
PCR recipe |
A |
B |
C |
|
dNTPs (1.25 mM each) |
1.0 |
1.0 |
1.0 |
|
MgCl22 (25 mM) |
1.0 |
1.0 |
0.5 |
|
10 X buffer * |
1.0 |
1.0 |
1.0 |
|
Forward Primer (5μM) |
0.6 |
0.6 |
1.0 |
|
Reverse Primer (5μM) |
0.6 |
0.6 |
1.0 |
|
Betaine (5 M) |
0 |
2.0 |
0 |
|
DMSO (100%) |
0 |
0.5 |
0 |
|
Taq polymerase* |
0.075 |
0.075 |
0.075 |
|
H2O |
2.725 |
0.225 |
2.425 |
|
DNA (10 ng/μL) |
3.0 |
3.0 |
3.0 |
|
Total |
10.0 |
10.0 |
10.0 |
* 10 X buffer is provided with purchase of Taq polymerase
Table 2. PCR programs for UMB SSR markers
|
Stage |
Temp. (°C) |
Time (min.) |
Cycles |
Stage |
Temp. (°C) |
Time (min.) |
Cycles |
|
|
|
||||||
|
Program A* |
Program F* |
||||||
|
Initial Hold |
95 |
|
1 |
Initial Hold |
- |
- |
- |
|
Thermal Cycles-1 |
94 |
|
18 |
Pre-PCR |
95 |
|
1 |
|
64 |
0:30 |
58 |
|
||||
|
72 |
|
72 |
|
||||
|
Thermal Cycles-2 |
94 |
|
30 |
Thermal Cycles |
94 |
0:30 |
30 |
|
55 |
|
58 |
0:30 |
||||
|
72 |
|
72 |
0:30 |
||||
|
Ending Hold |
72 |
|
1 |
Ending Hold |
72 |
|
1 |
|
Final Hold |
4 |
∞ |
1 |
Final Hold |
4 |
∞ |
1 |
|
|
|
|
|
|
|
|
|
|
Program J* |
Program L58 |
||||||
|
Initial Hold |
95 |
|
1 |
Initial Hold |
95 |
|
1 |
|
Thermal Cycles |
94 |
|
35 |
Thermal Cycles |
94 |
0:10 |
30 |
|
60 |
|
58 |
0:20 |
||||
|
72 |
|
72 |
0:45 |
||||
|
Ending Hold |
72 |
|
1 |
Ending Hold |
72 |
|
1 |
|
Final Hold |
4 |
∞ |
1 |
Final Hold |
4 |
∞ |
1 |
* Programs A, F and J are as in Ramsay et al., 2000.
Table 3. PCR and mapping information for UMB markers.
|
|
|
|
Population2 |
Quality |
|
PCR |
PCR |
|
|
Name |
Chr. |
BIN1 |
Polymorphism |
Mapped |
Score3 |
Alleles4 |
Program |
Recipe |
|
UMB101 |
1 (7H) |
11-12 |
A/M; F/S; H/O; H/L; S/M |
A/M; F/S; S/M |
2 |
3 |
L58 |
A |
|
UMB102 |
1 (7H) |
11-12 |
A/M; H/O; H/L; S/M |
S/M |
2.5 |
2 |
L58 |
A |
|
UMB103 |
1 (7H) |
13 |
A/M; F/S; H/O; H/L; S/M |
A/M; F/S; S/M |
2 |
4 |
L58 |
A |
|
UMB104 |
1 (7H) |
7-8 |
C/M; F/S; H/O; H/L; S/M |
F/S; S/M |
1.5 |
2 |
F |
A |
|
UMB105 |
1 (7H) |
10-11 |
A/M; C/M; H/O |
A/M; C/M |
3 |
2 |
L58 |
A |
|
UMB106 |
1 (7H) |
4-5 |
C/M; H/L; S/M |
C/M; S/M |
2 |
2 |
L58 |
A |
|
UMB107 |
1 (7H) |
4-5 |
F/S; H/O |
F/S |
3 |
4 |
J |
C |
|
UMB108 |
1 (7H) |
2 |
A/M; F/S; H/O; H/L; S/M |
F/S; S/M |
1 |
3 |
A |
B |
|
UMB201 |
2 (2H) |
6-7 |
C/M; F/S; H/O; H/L; S/M |
C/M; F/S; S/M |
2.5 |
4 |
L58 |
A |
|
UMB202 |
2 (2H) |
6-7 |
A/M; C/M; F/S; H/O; S/M |
A/M; C/M; F/S; S/M |
2 |
2 |
F |
A |
|
UMB203 |
2 (2H) |
4-5 |
C/M; H/L |
C/M |
2 |
2 |
J |
C |
|
UMB204 |
2 (2H) |
7-8 |
F/S; H/O |
F/S |
2.5 |
2 |
L58 |
A |
|
UMB205a |
2 (2H) |
4-5 |
C/M |
C/M |
1 |
2 |
J |
C |
|
UMB205b |
2 (2H) |
4-5 |
A/M; C/M; H/O |
C/M |
1 |
3 |
J |
C |
|
UMB206 |
2 (2H) |
2-3 |
C/M; F/S; H/O; H/L; S/M |
F/S; S/M |
2 |
3 |
L58 |
B |
|
UMB301 |
3 (3H) |
1-2 |
C/M; F/S; H/L; S/M |
C/M; F/S; S/M |
3 |
3 |
F |
A |
|
UMB302 |
3 (3H) |
4-6 |
A/M; C/M; F/S; H/O; S/M |
A/M; C/M; F/S; S/M |
1 |
4 |
L58 |
A |
|
UMB303 |
3 (3H) |
4-6 |
A/M; C/M; F/S; H/O; S/M |
A/M; C/M; F/S; S/M |
2 |
6 |
L58 |
A |
|
UMB304 |
3 (3H) |
4-6 |
A/M; C/M; F/S; H/L; S/M |
A/M; C/M; S/M |
1 |
4 |
J |
C |
|
UMB305 |
3 (3H) |
2-4 |
A/M; H/O; H/L |
A/M |
1 |
2 |
F |
A |
|
UMB306 |
3 (3H) |
2-4 |
A/M; H/O; H/L |
A/M |
1 |
3 |
F |
A |
|
UMB307 |
3 (3H) |
2-4 |
A/M; H/O; H/L |
A/M |
1 |
3 |
L58 |
A |
|
UMB308 |
3 (3H) |
1-2 |
A/M; C/M; F/S; H/O; S/M |
F/S; S/M |
2.5 |
2 |
F |
B |
|
UMB309 |
3 (3H) |
4-6 |
A/M; C/M; F/S; S/M |
S/M |
3 |
4 |
J |
B |
|
UMB310 |
3 (3H) |
4-6 |
A/M; C/M; F/S; H/L; S/M |
C/M; S/M |
2 |
3 |
L58 |
B |
|
UMB311 |
3 (3H) |
16 |
CM; F/S; H/L |
C/M |
3 |
3 |
J |
B |
|
UMB401 |
4 (4H) |
4-5 |
H/O; H/L; S/M |
S/M |
1 |
2 |
J |
B |
|
UMB402 |
4 (4H) |
2-3 |
H/O; H/L; S/M |
S/M |
1 |
2 |
L58 |
A |
Table 3 cont. PCR and mapping information for UMB markers.
|
|
|
|
Population2 |
Quality |
|
PCR |
PCR |
|
|
Name |
Chr. |
BIN1 |
Polymorphism |
Mapped |
Score3 |
Program |
Recipe |
|
|
UMB403 |
4 (4H) |
4-5 |
A/M; C/M; H/O; H/L; S/M |
C/M; S/M |
2 |
4 |
L58 |
B |
|
UMB404 |
4 (4H) |
5-6 |
C/M; H/O; S/M |
C/M; S/M |
1 |
2 |
J |
B |
|
UMB501a |
5 (1H) |
6 |
C/M; F/S |
C/M; F/S |
2 |
2 |
L58 |
B |
|
UMB501b |
5 (1H) |
6 |
C/M; F/S |
C/M; F/S |
3 |
2 |
L58 |
A |
|
UMB502 |
5 (1H) |
12-13 |
A/M; F/S; H/O |
A/M; F/S |
1 |
3 |
L58 |
A |
|
UMB503 |
5 (1H) |
2 |
A/M; C/M; F/S; H/O; H/L; S/M |
C/M; S/M |
2 |
6 |
L58 |
A |
|
UMB504 |
5 (1H) |
11-12 |
A/M; F/S; H/O; H/L; S/M |
F/S; S/M |
2 |
4 |
L58 |
B |
|
UMB505 |
5 (1H) |
2 |
A/M; C/M; H/O; H/L; S/M |
S/M |
3 |
3 |
J |
B |
|
UMB506 |
5 (1H) |
6 |
C/M; F/S; H/O; H/L |
C/M; F/S |
2 |
2 |
J |
B |
|
UMB507 |
5 (1H) |
6 |
C/M; F/S; H/O; H/L |
C/M; F/S |
1 |
3 |
L58 |
B |
|
UMB508 |
5 (1H) |
14 |
A/M; C/M; F/S; H/O; H/L |
C/M |
2.5 |
3 |
L58 |
B |
|
UMB601 |
6 (6H) |
13-14 |
A/M; F/S; S/M |
A/M; F/S; S/M |
1 |
2 |
F |
B |
|
UMB602 |
6 (6H) |
11-13 |
A/M; C/M; F/S; H/O; H/L |
A/M; C/M; F/S |
2 |
4 |
F |
A |
|
UMB603 |
6 (6H) |
13-14 |
A/M; C/M; F/S; H/O; H/L; S/M |
A/M; C/M; F/S; S/M |
2 |
4 |
L58 |
A |
|
UMB604 |
6 (6H) |
7-9 |
A/M; C/M; F/S; H/O; H/L; S/M |
C/M; F/S; S/M |
1 |
5 |
L58 |
A |
|
UMB605 |
6 (6H) |
14 |
A/M; C/M; F/S; H/O; H/L |
C/M; F/S |
1 |
2 |
L58 |
B |
|
UMB606 |
6(6H) |
1 |
A/M; C/M; F/S; H/O; H/L; S/M |
S/M |
3 |
3 |
F |
B |
|
UMB701 |
7 (5H) |
9 |
A/M; F/S; H/O |
F/S |
2 |
2 |
J |
B |
|
UMB702 |
7 (5H) |
10-11 |
C/M; H/O; H/L |
C/M |
1 |
3 |
F |
A |
|
UMB703 |
7 (5H) |
7-8 |
A/M; S/M |
S/M |
1.5 |
2 |
L58 |
A |
|
UMB704 |
7 (5H) |
2-4 |
A/M; F/S; H/O; H/L |
A/M; F/S |
1 |
4 |
L58 |
A |
|
UMB705 |
7 (5H) |
5-6 |
F/S; H/O |
F/S |
1.5 |
3 |
L58 |
A |
|
UMB706 |
7 (5H) |
9 |
A/M; C/M; H/O; H/L |
A/M; C/M |
1 |
2 |
L58 |
A |
|
UMB707 |
7 (5H) |
6-7 |
A/M; F/S; H/O; S/M |
A/M; S/M |
1 |
3 |
L58 |
A |
|
UMB708 |
7 (5H) |
10-11 |
A/M; H/O |
A/M |
1 |
2 |
F |
A |
|
UMB709 |
7 (5H) |
10-11 |
|
A/M; H/O; S/M |
S/M |
2 |
2 |
F |
|
UMB710 |
7 (5H) |
11-12 |
|
S/M |
S/M |
2 |
2 |
L58 |
|
UMB711 |
7 (5H) |
11-12 |
|
A/M; F/S; H/O; H/L; S/M |
F/S; S/M |
2 |
3 |
L58 |
Table 3 cont. PCR and mapping information for UMB markers.
|
|
|
|
|
Population2 |
Quality |
|
PCR |
PCR |
|
|
Name |
Chr. |
BIN1 |
|
Polymorphism |
Mapped |
Score3 |
Alleles4 |
Program |
Recipe |
|
UMB712 |
7 (5H) |
13-14 |
|
A/M; F/S; H/O; H/L; S/M |
F/S; S/M |
2 |
3 |
F |
B |
|
UMB713 |
7 (5H) |
13-14 |
|
A/M; F/S; H/O; S/M |
S/M |
2 |
4 |
L58 |
B |
|
UMB714 |
7 (5H) |
13-14 |
|
A/M; F/S; H/O; H/L; S/M |
F/S |
3 |
2 |
F |
B |
|
UMB715 |
7 (5H) |
13 |
|
A/M; F/S; H/O |
F/S |
2 |
5 |
J |
B |
|
UMB001 |
|
|
|
H/L |
|
2 |
3 |
L58 |
B |
|
UMB002 |
|
|
|
H/O |
|
3 |
2 |
F |
A |
|
UMB003 |
|
|
|
H/O |
|
2 |
4 |
F |
A |
|
UMB004 |
|
|
|
H/O; H/L |
|
3 |
2 |
J |
C |
|
UMB005 |
|
|
|
H/O |
|
1.5 |
2 |
L58 |
A |
|
UMB006 |
|
|
|
H/O |
|
3 |
2 |
L58 |
A |
|
UMB007 |
|
|
|
H/O; H/L |
|
2 |
3 |
L58 |
A |
|
UMB008 |
|
|
|
H/O; H/L |
|
1 |
3 |
A |
C |
|
UMB009 |
|
|
|
H/O |
|
2 |
3 |
L58 |
A |
|
UMB010 |
|
|
|
A/M; H/O; H/L; S/M |
|
2.5 |
3 |
F |
A |
|
UMB011 |
|
|
|
A/M; H/O |
|
1 |
3 |
L58 |
B |
|
UMB012 |
|
|
|
H/O |
|
1 |
2 |
L58 |
B |
|
UMB013 |
|
|
|
H/L |
|
2 |
2 |
L58 |
B |
|
UMB014 |
|
|
|
A/M; C/M; H/O; H/L |
|
2 |
3 |
J |
B |
|
UMB015 |
|
|
|
C/M; H/O; H/L; S/M |
|
3 |
5 |
F |
B |
|
UMB016 |
|
|
|
H/O |
|
1 |
2 |
A |
B |
1 Bold type indicates BINs with poor SSR marker coverage
2 Mapping populations: A/M=Atahualpa x M81; C/M=Chevron x M69; F/S=Frederickson x Stander;
H/O=Harrington x OUH602; H/L=Hor211 x Lacey; S/M=Steptoe x Morex
3 Scale from 1-5 where 1=Very easy to score and 5=Very hard to score
4 Number of Alleles based on the twelve mapping parents
Table 4. Design information for UMB primers.
|
|
EST |
Forward |
Reverse |
|
Name |
Accession1 |
Primer |
Primer |
|
UMB101 |
BE421034 |
CGGGTTCCATTGAGAAGAAC |
CACAAATACAGATGCCGCAC |
|
UMB102 |
CB881209 |
TTGTGTTTGAGATATCCTGTACTTTTC |
ACCTTTTGCCGGCTTTTATT |
|
UMB103 |
BQ762328 |
TGCCCATGAAGCCTCTTTAC |
GGAACGGAGGGAGTATTAAGC |
|
UMB104 |
CB881555 |
GGAAAAATAAACTATTCAACATCCTG |
CAGCGCATGTGTTCTCAGAT |
|
UMB105 |
BJ485220 |
GCCCCTGGTAAGAACTCCAT |
CTGGGAACCGTACAGTGTTG |
|
UMB106 |
AL502019 |
AGCATAAAGCCGCAAAAGAA |
GCGTCCTGATGAAGAGGTGT |
|
UMB107 |
CB873957 |
ACGCACGGGCATTTGTACT |
GCCTGCATCATTTGTTTGTG |
|
UMB108 |
CV055381 |
TCAAGCTGCTGCATTGCT |
AGCCCAAACCCTTTTGTTTT |
|
UMB201 |
BJ480735 |
GCTCCTGAAAAGGACCTCAG |
TCTCCGCCACCTACACATAG |
|
UMB202 |
BG299528 |
GGTCGGCTCCCTCTTCTACT |
CGAGCGACATGAGGAACAT |
|
UMB203 |
CB873608 |
TTTCATTGCTGTGACGGATG |
AGCCTCACCCGGACTACC |
|
UMB204 |
BM371159 |
GAATCCTCGGCCTTCTCAAC |
GCGGAGCTTGACCTCGAC |
|
UMB205a |
CB881957 |
CGGTCGTAGAACGGAATCAG |
GCACTTCCACCACAAGAAGC |
|
UMB205b |
CB881957 |
CGGTCGTAGAACGGAATCAG |
GCACTTCCACCACAAGAAGC |
|
UMB206 |
CB863325 |
GCGCTAGCTATCCACACAAA |
AACATTAAGGGCGACAAGGA |
|
UMB301 |
AV944239 |
CTTCACATGTCTGGGAAAACA |
GACATGTTGGAAGGTGGCTT |
|
UMB302 |
CD663662 |
ACCACAGGTAACCTCGCAAC |
AAAGTGCTGGGAGCTTGAAA |
|
UMB303 |
BU979287 |
CACGAGGGATGCTCTTGAGT |
TGTATATTTCAAGCTCCCAGCA |
|
UMB304 |
BM443659 |
CTTCGCTTACCGCTTTCG |
TTTCAAGCTCCCAGCACTTT |
|
UMB305 |
BJ484842 |
CAGAGCGGGCTCAACGTA |
ACTTGCTGTCATCCTTGCTG |
|
UMB306 |
BJ467519 |
GCAGAGCTGGCTCAACGTA |
TTCACTGATCGACCACTTGC |
|
UMB307 |
BJ461914 |
CTGCAGAGCTGGCTAACGTA |
TTCACTGATCGACCACTTGC |
|
UMB308 |
CB859861 |
CCCCTCAGGTTGTTCATCAT |
AGCAGCAGCAACAACAACAG |
|
UMB309 |
BF265771 |
GCTCGACTTCGAGGACACC |
ATTCTTGCGGAACGACCTC |
|
UMB310 |
CB879994 |
CTCCCAGCACTTTCACCATC |
CCGATGCTCTTGAGTCGTG |
|
UMB311 |
CA592691 |
ATCCAGTTTCAGCCACCAAC |
ACCGCAGTGATCAGTGACAA |
|
UMB401 |
AL507067 |
CGTCTTCGTACTCGCCTCTC |
ATCGAGATGCACTCCCTCAT |
|
UMB402 |
BQ466542 |
TCGATCCATCCAAACATGAG |
CGTGTCACGTGTGTGTGTGT |
Table 4 cont. Ordering and design information for UMB primers.
|
|
EST |
Forward |
Reverse |
|
Name |
Accession1 |
Primer |
Primer |
|
UMB403 |
BM372825 |
TTCCGCAGATTCATTTCCAC |
GCTGGACAGGCGTTAAAAAG |
|
UMB404 |
AJ475924 |
GGAGGCAAGAACACTTGACAG |
GCTCGATCTCCTCCTTGTTG |
|
UMB501a |
BE421033 |
CACACAGGCGACCATTTTC |
CAGCTAGACGCTATGAGCCA |
|
UMB501b |
BE421033 |
CACACAGGCGACCATTTTC |
CAGCTAGACGCTATGAGCCA |
|
UMB502 |
BI953342 |
ATCCCATCTCCCTCCTCCTA |
TGGAGTGCTCCTCCCAGTAG |
|
UMB503 |
BQ766039 |
TCCCGGTGCCATATACAAAT |
TTTGATGAAACGAAGGGAAA |
|
UMB504 |
CV054443 |
CAAAGTGCGCGTGAGAAATA |
AATCACCACCAGCTTCTTGG |
|
UMB505 |
BE195848 |
ATGTTGCAGCAGAGCAGTTG |
ATTGTTGGGGTTGTTCTTGC |
|
UMB506 |
CD663377 |
CTCTTCCGTGAAACGAAACC |
CGAGCAAGGACGTGGTAGAT |
|
UMB507 |
CD663377 |
ATGTTTCAACAGGCCATTCC |
CATGAAAACAGATGACGATGC |
|
UMB508 |
BF621983 |
GATTAAGGCGTCCAATTCCA |
TCGGGATGTGAAGAAGGAAC |
|
UMB601 |
BJ486149 |
AAATACCGTATGGAGGGTGCT |
CTACCCCTACGTCCGAGATG |
|
UMB602 |
CB877685 |
AGGAGTGGGTCTCAGGTTTG |
CAAGCAGATGCAACTACACCA |
|
UMB603 |
CA006980 |
ATGAAACATCGCGAACTGTG |
ACTGCAGTGAGGGAAGCTGT |
|
UMB604 |
CA004840 |
GAGCAATCCCCTCATCCAAT |
TCTTTGGTTTCCTCGTGTCC |
|
UMB605 |
CD053629 |
GAGGCTTGTTCCTCAGACCA |
ATGAGGAAGAGCGGGATCAG |
|
UMB606 |
CV056304 |
CGAGCAGCAGCAGATCGT |
CTCCTGCGCTTGGAGAAG |
|
UMB701 |
BE421177 |
ACGTCGTGGATCAACGTGTA |
TTACATTGCGCACAGCTAGG |
|
UMB702 |
BF265777 |
CAGCATCCATCAGCAATGAA |
CATGTTTGGCTTCTTCGTCC |
|
UMB703 |
CA025623 |
GCCGCCTCTTACTCTTTGC |
GGAGATGCCGAGGGACTT |
|
UMB704 |
AV942720 |
ATCCTCCAACGAGGCACATA |
GAGTCCATTTCACGGAGACC |
|
UMB705 |
CB876579 |
TGCTGAGACACACACACACC |
CGATGCACGAAAAGCTGTAG |
|
UMB706 |
CB875298 |
TCAACAGATGACGTGCATGA |
TCACACATTGAGGGAGGACA |
|
UMB707 |
CB876579 |
TGCTGAGACACACACACACC |
CGATGCACGAAAAGCTGTAG |
|
UMB708 |
BE421505 |
CTCCTCAGCTCTGGAATGGA |
GCGCATATACAAGCCAAACA |
|
UMB709 |
CV063649 |
ACGACAAGACTATGGCAGCA |
AAGGTTTCCTCAGCCTGTGA |
|
UMB710 |
CV063745 |
ACAGTTCCCAACTTCCAAGC |
CAGCACATCAGCCCGTACT |
|
UMB711 |
BJ553047 |
GCAACGACACGTCTAAACCA |
GTGGTGTGGTGTCCTTCCTC |
Table 4 cont. Design information for UMB primers.
|
|
EST |
Forward |
Reverse |
|
Name |
Accession1 |
Primer |
Primer |
|
UMB712 |
CB881537 |
CAAGAAGGAACGAAGCCAAG |
TGCTAGTTTTTCCGGCTGAT |
|
UMB713 |
CB881537 |
ATCAGCCGGAAAAACTAGCA |
CAAGCGAAGAGGAAGAGGAA |
|
UMB714 |
CB881537 |
GCAAGCAAATCACACTCTGG |
GCGTCATCTACGGCTGATTC |
|
UMB715 |
CB879524 |
CCGCGCCTAATTAACAAAAA |
AGCTGACTGCTGACCAACCT |
|
UMB001 |
BI949870 |
TTCTCCATGTTCGGCTTCTT |
ATCAGCAATGAAGTTGTCGG |
|
UMB002 |
BJ481660 |
GGCAAGTGAGCTCAACCTCT |
GCCCTCACATGCAACATCTA |
|
UMB003 |
CB881957 |
TCAGTGGTGACCGTGGTATC |
GCACTTCCACCACAAGAAGC |
|
UMB004 |
BG309433 |
ACCACGTCCACCACCATC |
CTCTTTCTCCGCTCATCACC |
|
UMB005 |
BQ763410 |
AAGAAAGCCCGGAAGAGAAG |
ATGGTGCCATCCTGATTGAT |
|
UMB006 |
BQ662872 |
CGGAGTCGCTTTCGAGATT |
ACTGACAGCAACGGTGGTAG |
|
UMB007 |
CB881851 |
CGCAATAAAATCGCAGGAAT |
ACGGAAGACCGGGATAGTTT |
|
UMB008 |
BU996005 |
CTGGAGCCTAGCTTGGAGTG |
CTCCACCGTTCTTCACGTTT |
|
UMB009 |
BI954579 |
CATCCCCATCCAGATCCA |
GTCGAGAGGTGCGAGGATT |
|
UMB010 |
BF621983 |
GCCACAGCCAAGAAAGCTAC |
GATGGGATCTGCTTGGAGAG |
|
UMB011 |
AV918630 |
TCTTCTCGCTAGCATCAGCA |
CAAAGAAGGAGGTGGCTTCA |
|
UMB012 |
BI960225 |
ATAGCGACGTGCTCCAGAGA |
AGCAGCGACTTCCTTAGCAG |
|
UMB013 |
CB873614 |
GAGCAAGCACGCACGTATTA |
GGGACCTCGAGATGATCAAG |
|
UMB014 |
CB882192 |
CAGGAGATCCGCGCTCTT |
CGAGCAGTGAACGATGTACG |
|
UMB015 |
CA030737 |
CGGACGAGGTTTACTCCAAA |
AGCACAGGAGGATGAGGATG |
|
UMB016 |
CA032410 |
CCATCACCCATTTCTTCCTC |
GATGGATTTGTCCGTCCAAG |
1 EST accession from which the primers were designed (http://www.ncbi.nih.gov/)
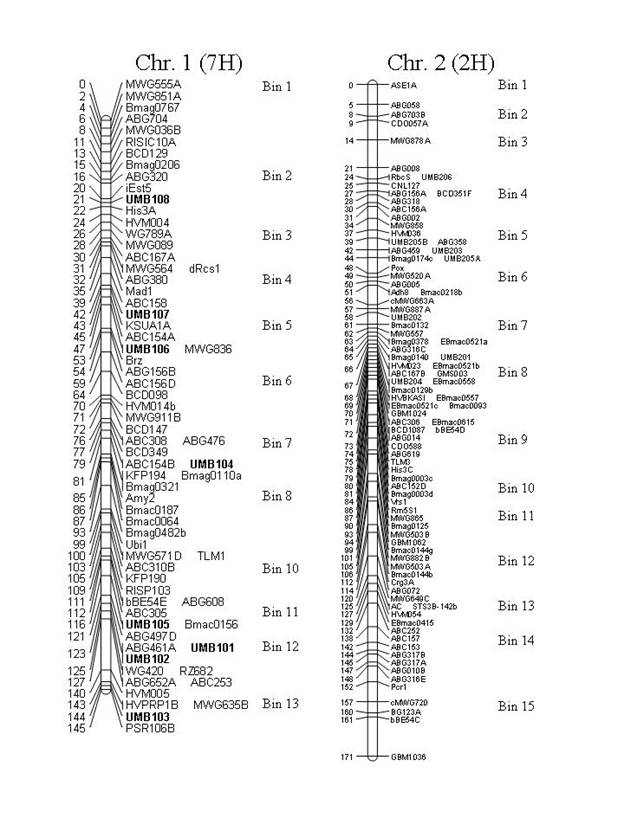
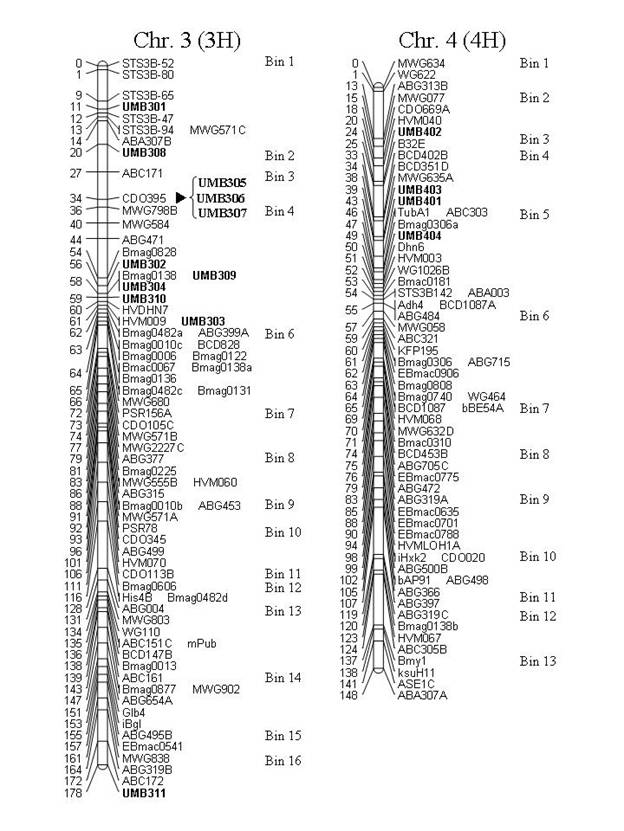
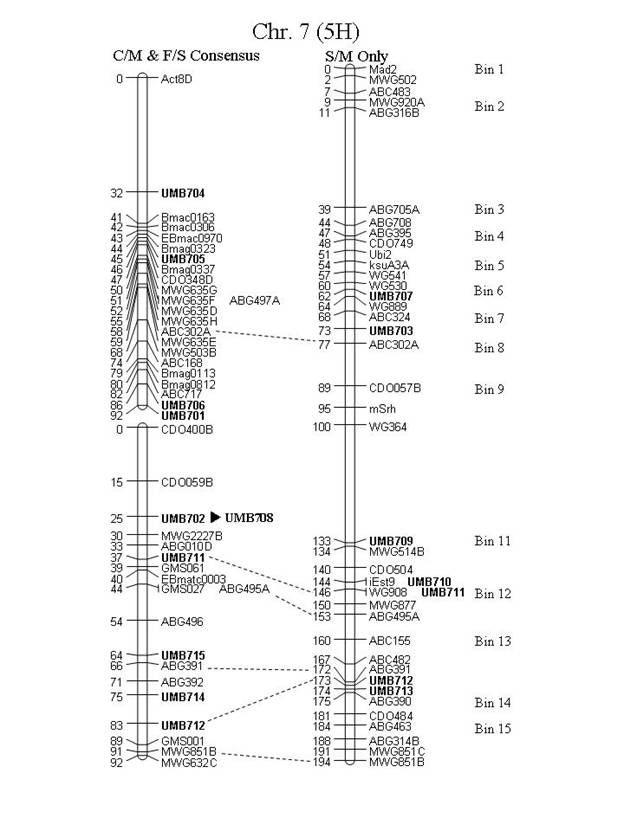
Figure 1. Map locations of the UMB markers on consensus
maps of the Steptoe x Morex (S/M), Frederickson x Stander (F/S) and Chevron x
M69 (C/M) populations. Chromosome 7 (5H)
is presented as a consensus map of the F/S and C/M populations with the S/M
population separate. UMB508 (denoted
with 'Ş') has had its location inferred from
the C/M population (C/M is included in the consensus but the inferred marker
did not map in the consensus). Markers
denoted with 'u' have had their location inferred from the
Atahualpa x M81 (A/M) population. Marker
"ABG497D" [chr. 1 (7H), BIN 11], was named "ABG497B" in
Canci et al. (2003), but the "ABG497B" locus should map to chr. 1 (7H),
BIN 4, therefore we have designated a new locus name.



