Barley Genetics Newsletter 36:12 - 16 (2006)
SSR Linkages to Eight Additional
Morphological Marker Traits
Lynn S. Dahleen and Jerome D. Franckowiak
USDA-Agricultural Research Service, PO Box 5677, SU Station, Fargo, ND 58105, and the Dept. Plant Sciences, North Dakota State Univ., Fargo, ND 58105
Introduction
More than 1000 morphological markers have been identified in barley (Franckowiak and Lundqvist 2002). These phenotypic traits were observed as spontaneous or artificially generated mutants in a wide range of cultivars over decades of research. Traits identified with alternate alleles include plant height, spike morphology, and seed size, among others. Approximately 275 of these markers have been placed on the morphological marker linkage map of the seven barley chromosomes (Franckowiak unpublished 2002) and/or on the consensus molecular marker linkage map (Kleinhofs 2002); approximately 150 to 200 additional markers have been placed on other maps but the rest have not been mapped.
Simple sequence repeat (SSR) markers are PCR-amplified regions of two or three base DNA repeats. Primers were designed to anneal to DNA on either side of each repeated segment, so size differences in amplification products are caused by different numbers of repeats in different genotypes. PCR reactions are easy to set up, do not use any hazardous chemicals, and only take a few hours for amplification. Products can be separated on agarose or acrylamide gels and results can be obtained in one day, which make these molecular markers ideal for short-term projects. We have worked with high school students in the North Dakota Governor’s School to map morphological traits over the last several summers. This report describes the location of eight additional morphological markers through linkage to SSR markers.
Materials and Methods
Mapping population development A
set of mutant lines were selected from various barley collections based on phenotype
and backcrossed four to six times to ‘Bowman’ (Table 1). Homozygous backcrossed
lines, BCnF2 populations, and BCnF2-derived
F3 lines were developed for mapping with molecular markers. Fifty BCnF2
seeds were sown for each mutant line in a greenhouse, along with the homozygous
mutant parent and Bowman. The greenhouse was maintained at 21-26oC
with a 16 h day/8 h night cycle supplied by sodium halide lights. Each plant
was scored as either normal or mutant. Plants were grown to maturity and
harvested. Twelve BCnF3 seed were sown for each BCnF2
plant to identify heterozygous lines. Parent and BCnF2 seed
were sown one per 15 cm clay pot, in a soil-less potting mix supplemented with
a slow release fertilizer (14-14-14). The BCnF3 seed were
sown with six seed per 15 cm clay pot and scored for the mutant trait at the
appropriate time. Plants were treated with
Trait Analysis Backcross-derived
near-isogenic lines (NILs) and Bowman were planted in a field near
Molecular marker testing Leaf tissue was harvested from young parent and BCnF2 plants grown in the greenhouse and the DNA extracted using the method of Dahleen et al. (2003). The DNA was then resuspended in 200 mL of modified TE (10 mM Tris-Cl, pH 7.4 and 0.1 mM EDTA). Simple sequence repeat (SSR) markers (Ramsay et al. 2000) were screened against the mutant near-isogenic lines and Bowman to identify polymorphisms. The PCR methods used were described in Dahleen et al. (2003). Amplified fragments were separated by gel electrophoresis using 4% Super Fine Resolution (SFR) agarose (Amresco, Solon, OH) in1 X TAE (40 mM Tris-acetate and 1 mM EDTA). Alternatively, acrylamide gel electrophoresis was used as described by Wang et al. (2003). The gels were stained with ethidium bromide, and photographed under UV light. Markers that detected polymorphisms between Bowman and a mutant near isogenic line were tested on the corresponding BCnF2 population. SSR and mutant trait segregation data were entered into MAPMAKER software (Lander et al. 1987; Lincoln et al. 1992) to test linkage between the markers and the mutant trait.
|
Table 1. Morphological markers, Barley Genetic Stock number, their chromosome locations, pedigrees of the mapping populations and the number of backcrosses to Bowman to develop the near-isogenic lines |
||||
|
Gene |
BGS No. |
Chromosome |
Pedigree |
Backcrosses |
|
cer-zt.389 |
BGS437 |
2H |
cer-zt.389/5*Bowman |
4 |
|
dsp.ah |
|
7H |
DWS1180 Mut4841/6*Bowman |
5 |
|
dsp.at |
|
3H |
Bowman*5/DWS1220 7117 |
4 |
|
dsp.ba |
|
7H |
DWS1357 UT1713/6*Bowman |
5 |
|
int-k.47 |
BGS546 |
7H |
int-k.47/7*Bowman |
6 |
|
nec.50 (pmr2) |
BGS634 |
7H |
Bowman*5/nec.50 |
4 |
|
nec.54 (pmr2) |
BGS634 |
7H |
Bowman*5/nec.54 |
4 |
|
pyr.ai |
|
3H |
Bowman*6/DWS1018 |
5 |
Results and Discussion
Linkage between SSR markers and the morphological traits was identified for each of the eight backcrossed morphological marker lines. All F2 populations segregated 3:1 normal:mutant as expected for single recessive traits. There were no significant differences between Bowman and near-isogenic lines for lodging, leaf width or heading date.
The eceriferum
mutant cer-zt.389 (BGS 437) was
located approximately in bin 1 of chromosome 2H, linked to Bmac0134 (Fig. 1).
This NIL only differed from Bowman for kernels per spike, with a significant
increase in cer-zt. The intermedium
spike mutant int-k.47 (BGS 546) was
located in the centromeric region of chromosome 7H, closely linked to markers
Bmag0217 and Bmac0162 in bins 6 to 7. The int-k.47
NIL had significantly reduced height, peduncle length, awn length,
kernels/spike, leaf length, kernel weight, and yield. The two necroticans
mutants (nec.50 and nec.54) are alleles at the premature
ripe 2 (pmr2) locus (BGS 634).
Analysis of individual populations indicated both were linked to the same
markers on chromosome 7H (Fig. 1). Because they are alleles, a combined linkage
analysis was conducted, which showed tight linkage to markers in bin 5 of 7H
(Fig. 2). The nec.54 NIL was analyzed
in the field and showed reduced yield compared to Bowman. The pyramidatum
mutant pyr.ai was linked between
markers on chromosome 3H in bins
Three additional dense spike mutants were mapped, dsp.ah, dsp.at, and dsp.ba. The first two were linked to markers on chromosome 7H near the centromere while dsp.ba was located near the centromere on the short arm of chromosome 3H (Fig. 1). Previously located dense spike loci include dsp1 on chromosome 7HS, dsp9 on chromosome 6HL, and dsp10 on chromosome 3HS. Agronomic trait performance of these three lines was similar to Bowman for most traits. The dsp.ah mutant showed significantly reduced height, peduncle length and rachis internode length, and lower yields but showed a significant increase in the number of kernels per spike. The dsp.at mutant was shorter and had reduced peduncle and rachis internode lengths, while dsp.ba only showed a decrease in rachis internode length compared to Bowman. These differences in the dsp mutants indicate it is unlikely they are alleles, and they are unlikely to be alleles of dsp1, ert-a, ert-d or ert-m, mutants with similar dense spikes.
In summary, we have placed eight additional morphological mutants on the barley molecular linkage map. One (cer-zt) mapped to chromosome 2H, two (dsp.ba and pyr.ai) mapped to chromosome 3H, and five (dsp.ah, dsp.at, int-k.47, nec.50 and nec.54) mapped to chromosome 7H.


Figure 1. Linkage maps showing positions of morphological markers in relation to SSR markers mapped in segregating populations of 50 F2 plants for each trait. Cumulative linkage distances are on the left of the vertical bar and marker names are on the right. Approximate chromosomal bin locations for the SSRs are in parentheses.
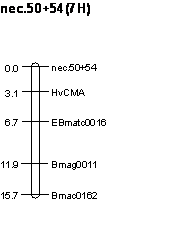
Figure 2. Linkage map showing the position of the nec.50 and nec.54 when the trait and marker data were combined for these alleles at the pmr2 locus (BGS 634).
References
Dahleen LS, Franckowiak JD and Vander Wal LJ. 2003. Exposing students and teachers to plant molecular genetics with short-term barley gene mapping projects. J Natural Resour Life Sci Educ 32:61-64.
Franckowiak JD and
Kleinhofs A. 2002. Integrating molecular and
morphological/physiological marker maps [Online]. Available at
http://wheat.pw.usda.gov/ggpages/bgn/32/index.html (verified
Lander E, Green P, Abrahamson J, Barlow A, Daley M, Lincoln S, and Newburg L. 1987. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174-181.
Lincoln
S, Daly M and Lander E. 1992. Constructing genetic maps with MAPMAKER/EXP 3.0.
Whitehead Inst. Tech. Rep. 3rd ed. Whitehead Inst.,
Ramsay L, Macaulay M, Ivanissevich S, MacLean K, Cardle L, Fuller J, Edwards KJ. Tuvesson S, Morgante M, Massari A, Maestri E, Marmiroli N, Sjakste T, Ganal M, Powell W, and Waugh R. 2000. A simple sequence repeat-based linkage map of barley. Genetics 156:1997-2005.
Wang D, Shi J, Carlson SR, Cregan PB, Ward RW, and Diers BW. 2003. A low-cost, high-throughput polyacrylamide gel electrophoresis system for genotyping with microsatellite DNA markers. Crop Sci 43:1828-1832.