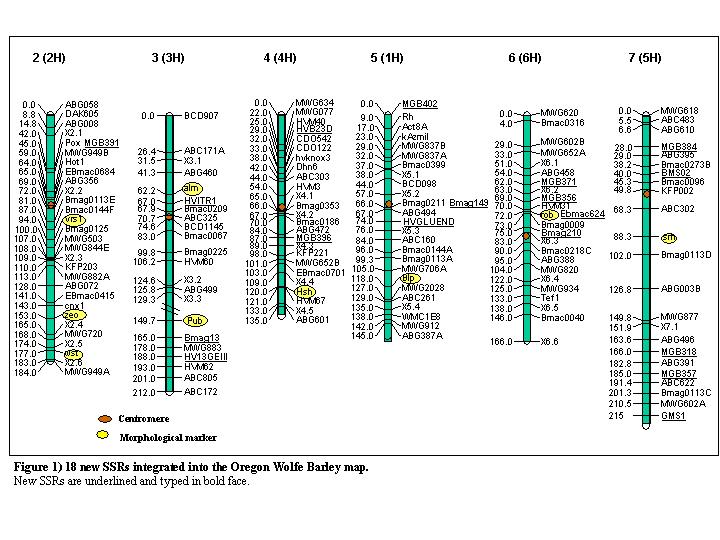
Insertion of 18 new SSR markers into the Oregon Wolfe Barley map
Maria von Korff, Judith Plümpe, Wolfgang Michalek1), Jens Léon, and Klaus Pillen
Department of Crop Science and Plant Breeding, University of Bonn, Katzenburgweg 5, 53115 Bonn, Germany, email. K.Pillen@uni-bonn.de
1)PLANTA GmbH, Grimsehlstr. 31, D-37555 Einbeck, Germany
Introduction
The Oregon Wolfe barley map is based on a population consisting of 94 doubled-haploid barley plants (Costa et al. 2001). The OWB population integrates molecular and morphological marker loci, shows a high level of polymorphism, and serves as a genetic teaching tool (http://barleyworld.org/). So far, thirteen morphological markers and several hundred DNA-based markers have been localized in the OWB linkage map. The majority of them are AFLP markers. The genetic map spans 1.387 cM with an average density of one marker every 1.9 cM. Here, we report on a further saturation of the OWB map with SSR markers. In total, 18 SSRs including eight novel EST-derived SSR markers were added to the map. References and primer sequences for each marker are given in Table 1.
Microsatellite markers
A total of 22 SSRs were screened for polymorphism between the OWB parents, Dom and Rec. Of these SSRs, 18 revealed a polymorphism in the OWB population. Eight EST-derived SSR markers, abbreviated with “MGB”, were newly developed from EST sequences (Michalek et al. 2002) according to Pillen et al. (2000). In addition, one, one, three and five SSRs, previously published by Russell et al. (1997), Struss and Plieske (1998), Pillen et al. (2000) and Ramsay et al. (2000), respectively, were also mapped in OWB. In total 15 SSRs are mapped for the first time. These include all SSRs in Table 1 except Bmag210 (Perovic et al. 2000), Bmag13 (Ramsay et al. 2000) and GMS1 (Pillen et al. 2000).
SSRs were amplified with a standard PCR protocol using a touchdown annealing temperature of 64-55 °C (Pillen et al. 2000). The genotype data were analysed together with the OWB mapping data using the mapping program JoinMap (Stam 1993). The SSRs were integrated into the OWB map (http://barleyworld.org/) according to the results obtained with JoinMap (Fig.1).
Table 1) List of 18 SSR markers placed on the OWB map
|
Name |
Chr |
Pos (cM) |
EMBL accession |
Repeat |
Pri-mer |
Sequence |
Ref. a) |
|
MGB391 |
2 (2H) |
45 |
HY02P09T3 |
(CCG)4 |
F |
AGCTCCTTTCCTCCCTTCC |
1 |
|
|
|
|
|
|
R |
CCAACATCTCCTCCTCCTGA |
|
|
HVITR1 |
3 (3H) |
67 |
X65875 |
(G)12 |
F |
CCACTTGCCAAACACTAGACCC |
2 |
|
|
|
|
|
|
R |
ATTCATGCAGATCGGGCCAC |
|
|
Bmag13 |
3 (3H) |
165 |
|
(CT)21 |
F |
AAGGGGAATCAAAATGGGAG |
3 |
|
|
|
|
|
|
R |
TCGAATAGGTCTCCGAAGAAA |
|
|
HV13GEIII |
3 (3H) |
188 |
X67099 |
(AT)5 |
F |
AGGAACCCTACGCCTTACGAG |
2 |
|
|
|
|
|
|
R |
AGGACCGAGAGTGGTGGTGG |
|
|
HVB23D |
4 (4H) |
29 |
X76603 |
(GT)6 |
F |
GGTAGCAGACCGATGGATGT |
3 |
|
|
|
|
|
|
R |
ACTCTGACACGCACGAACAC |
|
|
MGB396 |
4 (4H) |
87 |
HY03C23T3 |
(CTT)6 |
F |
CGCTAGCTTGTTTCTCGTTTG |
1 |
|
|
|
|
|
|
R |
TCGCATGGCATCAACTACAG |
|
|
MGB402 |
5 (1H) |
0 |
HY03I05T3 |
(CCT)5 |
F |
CAAGCAAGCAAGCAGAGAGA |
1 |
|
|
|
|
|
|
R |
AACTTGTGGCTCTGCGACTC |
|
|
Bmag149 |
5 (1H) |
66 |
|
(CT)5TT(CT)10 |
F |
CAAGCCAACAGGGTAGTC |
3 |
|
|
|
|
|
|
R |
ATTCGGTTTCTAGAGGAAGAA |
|
|
HVGLUEND |
5 (1H) |
74 |
X56260 |
(T)11 |
F |
TTCGCCTCCATCCCACAAAG |
2 |
|
|
|
|
|
|
R |
GCAGAACGAAAGCGACATGC |
|
|
MGB371 |
6 (6H) |
62 |
HW02M04T3 |
(TG)6 |
F |
CACCAAGTTCACCTCGTCCT |
1 |
|
|
|
|
|
|
R |
TTATTCAGGCAGCACCATTG |
|
|
MGB356 |
6 (6H) |
69 |
HW01M13u |
(CTG)5 |
F |
TGGTCTGGAGCTCTCAACAG |
1 |
|
|
|
|
|
|
R |
AAGCCACATTGAAGGAGCAC |
|
|
EBmac624 |
6 (6H) |
72 |
|
(GT)6,(GT)7,(GT)7 |
F |
AAAAGCATTCAACTTCATAAGA |
3 |
|
|
|
|
|
|
R |
CAACGCCATCACGTAATA |
|
|
Bmag210 |
6 (6H) |
75 |
|
(AG)7T(AG)13 |
F |
ACCTACAGTTCAATAGCTAGTACC |
3 |
|
|
|
|
|
|
R |
GCACAAAACGATTACATCATA |
|
|
MGB384 |
7 (5H) |
28 |
HY02J05T3 |
(AGG)6 |
F |
CTGCTGTTGCTGTTGTCGTT |
1 |
|
|
|
|
|
|
R |
ACTCGGGGTCCTTGAGTATG |
|
|
BMS02 |
7 (5H) |
40 |
|
(CA)21 |
F |
AGAGTAGTGGAAAGAAAGTT |
4 |
|
|
|
|
|
|
R |
TGGTAGTGAGATGAGGTGAC |
|
|
MGB318 |
7 (5H) |
166 |
HK03N02R |
(CTG)6 |
F |
CGGCTCAAGGTCTCTTCTTC |
1 |
|
|
|
|
|
|
R |
TATCTCAGATGCCCCTTTCC |
|
|
MGB357 |
7 (5H) |
185 |
HW01M22T3 |
(CCG)8 |
F |
GCTCCAGGGCTCCTCTTC |
1 |
|
|
|
|
|
|
R |
AGCTCTCTCTGCACGTCCTT |
|
|
GMS1 |
7 (5H) |
215 |
|
(CT)7TTT(CT)2 |
F |
ctgaccctttgcttaacatgc |
5 |
|
|
|
|
|
|
R |
tcagcgtgacaaacaataaagg |
|
a) 1, new MGB-SSRs, derived from barley ESTs (Michalek et al. 2002); 2, Pillen et al. (2000) ; 3, Ramsay et al. (2000) ; 4, Russell et al. (1997) ; 5, Struss and Plieske (1998)
References
Costa J.M., Corey A., Hayes P.M., Jobet C., Kleinhofs A., Kopisch-Obusch A., Kramer S.F., Kudrna D., Li M., Riera-Lizarazu O., Sato K., Szucs P., Toojinda T., Vales M.I., and R.I. Wolfe. 2001. Molecular mapping of the Oregon Wolfe Barleys: an exceptionally polymorphic doubled-haploid population. Theor. Appl. Genet. 103:415-424.
Michalek W., Weschke W., Pleissner K.-P., and A. Graner. 2002. EST analysis in barley defines a unigene set comprising 4,000 genes. Theor. Appl. Genet. 104:97-103.
Perovic D., Smilde W.D., Haluskova J., Waugh R., Sasaki T., and A. Graner. 2000. Update of the Igri/Franka molecular marker map. Barley Genetics Newsletter 30:15-20.
Pillen K., Binder A., Kreuzkam B., Ramsay L., Waugh R., Förster J., and J. Léon. 2000. Mapping new EMBL-derived barley microsatellites and their use in differentiating German barley cultivars. Theor. Appl. Genet. 101:652-660.
Ramsay L., Macaulay M., degli Ivanissevich S., Maclean K., Cardle L., Fuller J., Edwards K. J., Tuvesson S., Morgante M., Massari A., Maestri E., Marmiroli N., Sjakste T., Ganal M., Powell W., and R. Waugh. 2000. A simple sequence repeat-based linkage map of barley. Genetics156:1997-2005.
Russell J., Fuller J., Young G., Thomas B., Taramino G., Macaulay M., Waugh R., and W. Powell. 1997. Discriminating between barley genotypes using microsatellite markers. Genome40:442-450.
Stam P. 1993. Construction of integrated genetic linkage maps by means of a new computer package: JoinMap. Plant Journal 3:739-744.
Struss D., and J. Plieske. 1998. The use of microsatellite markers for detection of genetic diversity in barley populations. Theor. Appl. Genet.97: 308-315.