TARGETED
SATURATION MAPPING OF A HIGH RECOMBINATION REGION IN BARLEY
USING ESTs
IDENTIFIED VIA SYNTENY TO RICE
David A. Kudrna1, Kara Johnson1
, Robert Brueggeman1 , Arnis Druka1, and Andris Kleinhofs2
1Department of Crop and Soil Sciences, Washington State
University,
Pullman, WA 99164-6420 USA
2Department of Crop and Soil Sciences and School of Molecular
Biosciences,
Washington State University, Pullman, WA
99164-6420 USA
Introduction
Physical mapping identified high, medium
and low recombination regions in the barley genome (Kunzel et al, 2000). On the hypothesis that recombination occurs
primarily in genes, the high and medium recombination regions are presumed to
be gene rich, whereas the low recombination regions are presumed to be
deficient or very low in gene sequences (Barakat, 1997). In order to validate Kunzel's map, we are
developing a Bacterial Artificial Chromosome
(BAC) based physical map for the barley chromosome 1(7H)S region defined by
markers cMWG703 and MWG836. This
chromosome region encompasses approximately 40 cM (Igri x Franka map) and was
projected to have a physical/genetic distance ratio of 100 kb per cM (Kunzel et
al., 2000).
A physical BAC clone contig could be, in
principle, developed by chromosome walking.
However, this is a very difficult undertaking and perhaps not possible
for a large chromosome region in a large genome organism such as barley. The alternative is saturation mapping
leading to "chromosome landing", a term coined by Tanksley et al.
(1995) to define an approach based on finding markers close enough to the gene
of interest to reside on the same large insert clone as the gene of
interest. Saturation mapping can be practiced
with random markers mapped in the whole genome. An example of this approach is whole genome mapping with AFLP
markers (Vos et al., 1995). This
technique, although very powerful, requires mapping very large numbers of
markers in order to have a statistical chance of finding a marker near the
target, particularly in a large genome.
A refinement of this approach is to target random markers at a specific
chromosome region using bulked segregant analysis (Michelmore et al., 1991) or
deletion lines to define the target.
Examples of such approaches can be found in Xu and Korban, (2000) and
Gill et al. (1996). The ideal would be
to target specific markers to a specific chromosome region. This was accomplished by Monna et al. (1997)
using yeast artificial chromosome (YAC) clones to saturate the region
containing the rice blast disease resistance gene Pi-b. However, barley, like most species, does not
have the luxury of a YAC clone physical map.
Previously we targeted specific rice markers to a specific barley genome
region exploiting barley-rice synteny (Kilian et al., 1995). This approach, while successful was still
not good enough to saturate the specific barley genome region with markers
close enough to allow "chromosome landing" with the ca. 100 kb insert
BAC library available for barley (Yu et al., 2000). Another approach is to exploit comparative mapping among many
species to target specific markers to a specific chromosome region. An example is the work of Faris et al.
(2000). Similar to the rice synteny,
this approach works, but is not sufficient for complete saturation. Here we describe an improvement of the rice
synteny method where we use the rice genomic sequence to identify EST clones
from barley and wheat for mapping to the region of interest. Using this method, we have saturated the
chromosome 1(7H)S target region between markers cMWG703 and MWG836 with markers
and identified multiple BAC contigs.
The work is in progress, but we expect that eventually we will have a
complete or nearly complete BAC contig of the region of interest. This optimism is based on our work showing
that the average barley BAC clone from the high or medium recombination regions
contains 4-5 genes (Rostoks et al., 2002).
Assuming that the Triticeae (barley and wheat) EST database contains
approximately 50% of all the genes, we should be able to find 2 markers per BAC
clone, on the average. There are,
however, some BAC clones that contain only one gene. Thus, some gaps may occur.
The limitations of this technique are only the available rice genomic
sequence and the number of deposited Triticeae EST sequences.
Methods
The
targeted gene rich region of barley chromosome 1(7H)S is flanked by the markers
cMWG703 and MWG836. Barley EST probes
were selected from the syntenic rice chromosome 6 region (molecular markers
C425A to S1434) by BLASTN analysis of the rice genomic sequences
(http://rgp.dna.affrc.go.jp/) against the barely EST database
(http://www.ncbi.nlm.nih.gov/BLAST/).
EST clones were selected as homologous to rice sequence based upon S-values
greater than 70 and E-values less than 1e-10.
EST probes are available from Clemson University Genome Institute (CUGI;
http://www.genome.clemson.edu/). Probes
were genetically mapped by RFLP using a set of recombinant lines selected from
the North American Barley Genome Project Steptoe x Morex doubled haploid
mapping population. Protein function
predictions were determined by BLASTX analysis of translated EST sequences with
the protein sequence database (http://www.ncbi.nlm.nih.gov/BLAST/).
Physical BAC contigs were assembled
manually following individual BAC confirmation with each EST. BAC clones and screening filters are
available from CUGI.
Results
Rice chromosome 6 from 6cM to 15 cM was
identified as the barley chromosome 1(7H)S 12cM to 38 cM (cMWG703-MWG836)
syntenic region by mapping rice probes in barley and barley probes in rice
(data not shown). Fortunately this rice
region has been nearly completely sequenced with just one gap.
A total of 106 loci have been mapped to
the chromosome 1(7H)S cMWG703 to MWG836 target region (Fig. 1). Of these, 59 are ESTs (Table 1) that were
identified by searching the barley EST database with rice PAC or BAC clone
genomic sequences. One is a disease
resistance locus (Rcs5) and another
is a morphological locus (Lga). The remaining 47 loci were mapped previously
using probes identified by synteny to wheat, rice, and oat maps or by random
mapping.
The sequence from 19 rice PAC or BAC
clones (there is considerable overlap in the sequence) from this region was
used to identify a total of 159 EST clones from the barley EST data base. Of these, 107 were genetically and/or
physically mapped to 118 loci; 59 loci in the HRR. Nine were non-polymorphic and 21 are still in process. Many of the EST clones were single copy, 17
are duplicated and 3 hybridized to multiple (more than 10) bands on genomic
Southern blots. Additionally, 2
retroelement-like ESTs were identified by homology to the rice sequence.
Fifty-seven EST loci mapped outside the
HRR (Table 2). There was no obvious
genetic clustering of these loci that would suggest a duplicated region in
barley, however, the number of probes mapping outside the target region is
higher than anticipated. There may be
several trivial explanations for this, other than chromosome rearrangements,
and we are still investigating this.
A total of 1149 BAC clones were
identified by hybridizing the 159 identified EST clones to the barley cv. Morex
6.3X BAC library. This identified 40 BAC
contigs, but 32 singletons (no overlap with BAC clones from other probes) still
remain (Fig.1). A crude estimate of the
physical size of the region covered with BAC clones to date yielded 5.5 Mb. This estimate was derived from the 100 kb
average size of the BAC clone inserts and by assuming 50% overlap for the
clones in contigs. Thus, the region is
already somewhat larger than the approximately 4 Mb (40 cM x 0.1 Mb per cM)
estimated by Kunzel et al. (2000).
However, given the translocation breakpoint method used and the lack of
infinite breakpoints, the Kunzel et al. estimate and our data are probably
within reasonable agreement. BAC
addresses identified by EST hybridization are available from our BAC database: http://barleygenomics.wsu.edu/db3/db3.html.
Discussion
Using the rice-barley EST homology
method, we have rapidly and efficiently more than doubled the saturation of the
target region from 50 to 110 markers.
The work described here was completed at a time when only about 65,000
barley EST sequences were available in the database. As of the end of Jan. 2002, a total of 144,000 barley and 73,000
wheat EST sequences have been deposited.
Thus, a new BLAST search of the Triticeae (barley and wheat) EST
database should result in an even higher level of marker saturation. To date we have not used the wheat EST
clones, but they should work as well as the barley EST clones. In addition, completion of the rice genomic
sequencing of the syntenous region will facilitate this work.
Our data, showing that approximately 50%
of the EST clones identified by the described procedure map to the target
region, suggests that it is a valid and rapid technique for saturation mapping
of specific chromosome regions in barley.
However, the number of probes mapping outside of the target region is
larger than anticipated and bothersome.
Some possible trivial explanations include selection of EST with too low
homology score, mapping of a related but not orthologous band, picking of the
wrong EST, and library contamination.
We are investigating these ideas to determine if the ratio of those
mapping to the right region vs. those mapping elsewhere can be improved.
The physical-genetic distance ratios are
highly variable in the barley genome.
Kunzel et al. (2000) has identified the major features of the genome,
but the variation within those features has remained hidden to the global
look. Thus, even in a high
recombination region we can expect to find substantial variation. We have observed that in this region. However,
to date we have observed very few BAC clones that span recombination
breakpoints.
The BAC contig of the target region, once
completed, will provide access to numerous interesting genes, including
seedling spot blotch resistance Rcs5,
leaf wax (Cer-ze, Gsh3), long glume
awn (Lga), nitrate assimilation (Nar3), winding dwarf (wnd), the chlorophyll genes Fch5, Yvs, possibly albino seedling 7 (abo7) and anthocyanin-less 1 (ant1).
Several QTL have also been mapped to this region including Fusarium head
blight, heading date, yield, net blotch resistance (adult), spot blotch
resistance (adult), tiller number, short and long day length, plant height,
plant grain weight, kernel length and shape, and crossability with wheat.
References
Barakat A., N.
Carels and G. Bernardi. 1997. The distribution of genes in the genomes of
Gramineae. Proc Natl Acad Sci USA 94: 6857-6861.
Faris, J.D.,
K.M. Haen and B.S. Gill. 2000. Saturation Mapping of a gene-rich
recombination hot spot region in wheat.
Genetics 154:823-835.
Gill, K. S.,
B. S. Gill, T. R. Endo and E. V. Boyko. 1996. Identification and high-density
mapping of gene-rich regions in chromosome group 1 of wheat. Genetics 144:
1883-1891
Kilian, A., D. A.
Kudrna, A. Kleinhofs, M. Yano, N. Kurata, B. Steffenson and T. Sasaki. 1995.
Rice-barley synteny and its application to saturation mapping of the
barley Rpg1 region. Nucleic Acids
Research 23:2729-2733.
Kunzel, G., L.
Korzum and A. Meister. 2000. Cytologically integrated physical restriction
fragment length polymorphism maps for the barley genome based on translocation
breakpoints. Genetics 154: 397-412.
Michelmore R.
W., I. Paran and R. V. Kesseli.
1991. Identification of markers
linked to disease-resistance genes by bulked segregant analysis: A rapid method
to detect markers in specific genomic regions by using segregating
populations. Proc. Natl. Acad. Sci. USA
88:9828-9832.
Monna,
L., A. Miyao, H.S. Zhong, M. Yano, M. Iwamoto, Y. Umehara, N. Kurata, H.
Hayaska and T. Sasaki. 1997. Saturation mapping with subclones of
YACs: DNA marker production targeting
the rice blast disease resistance gene, Pi-b.
Theor. Appl. Genet. 94:170-176.
Rostoks, N.,
Y-J Park, W. Ramakrishna, J. Ma, A. Druka, B.A. Shiloff, PJ. SanMiguel, Z.
Jiang, R. Brueggeman, D. Sandhu, K. Gill, J.L. Bennetzen, and A.
Kleinhofs. 2002. Genomic sequencing
reveals gene content, genomic organization and recombination relationships in
barely. Functional and Integrative
Genomics, in press.
Tanksley,
S.D., M.W. Ganal, and G.B. Martin, G.B. 1995.
Chromosome landing: a paradigm for map-based gene cloning in plants with
large genomes. Trends in Genetics
11:63-68.
Vos, P., R.,
Hogers, M., Bleeker, M. Reijans, T. van de Lee, M. Hornes, A. Frijters, J. Pot, J. Peleman, M. Kuiper and M.
Zabeau. 1995. AFLP: a new technique for DNA
fingerprinting. Nucleic Acids Research
23:4407-4414.
Xu, M.L. and
S.S. Korban. 2000. Saturation mapping
of the apple scab resistance gene Vf using AFLP markers. Theor. Appl. Genet. 101:844-851.
Yu, Y., J. P.
Tomkins, R. Waugh, D. A. Frisch, D. Kudrna, A. Kleinhofs, R. S. Brueggeman, G.
J. Muehlbauer, R. P. Wise, and R. A. Wing.
2000. A bacterial artificial
chromosome library for barley (Hordeum vulgare L.) and the identification of
clones containing putative resistance genes.
Theor Appl Genet 101: 1093-1099.
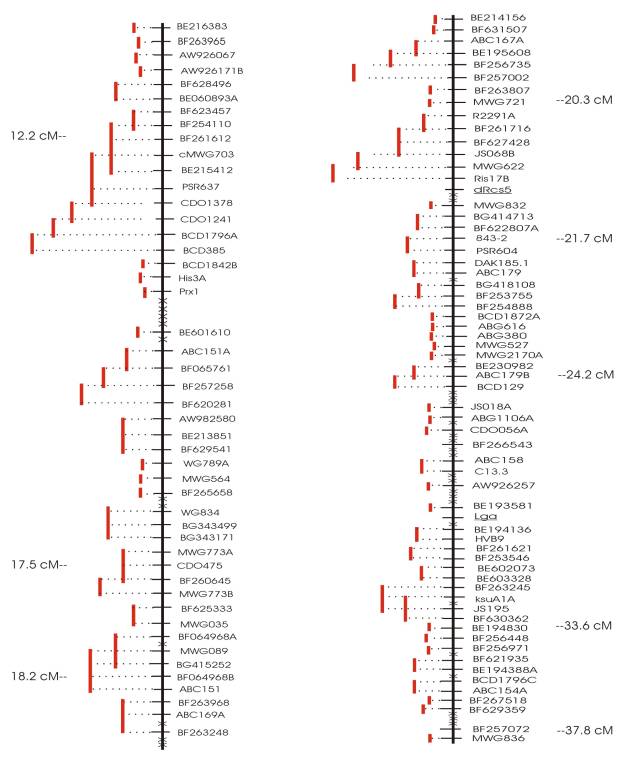
Figure
1
Click here for a .pdf file of Figure 1.
Genetic
and Physical maps of the high-recombination region of barley chromosome 1 (7H)S
between markers cMWG703 (bin 2) and MWG836 (bin 5). Genetic distances are shown in centimorgans (cM).
Table 1. Barley EST loci physically and/or
genetically mapped in the high-recombination region of barley chromosome 1(7H)S
between markers cMWG703 (bin 2) and MWG836 (bin 5).
|
(Acc.#) |
(Acc. #) |
EST-PAC S-value |
EST-PAC
E-value |
Predicted Gene Function |
Function S -value |
Function E -value |
|
BE230982 |
AB026295 |
SR |
SR |
proline-rich cell wall protein, sequence removed |
SR |
SR |
|
BE060893 |
AP002542 |
206 |
2e-50 |
40S ribosomal protein S20, rice |
218 |
2e-56 |
|
AW926257 |
AB026295 |
SR |
SR |
soluble starch synthase I, sequence removed |
SR |
SR |
|
BF253755 |
AP000391 |
109 |
3e-21 |
Beta-5 subunit of 20S proteasome, rice |
266 |
7e-71 |
|
BF256735 |
AP000399 |
98 |
2e-17 |
wak 1 gene, rice |
160 |
2e-38 |
|
BF257072 |
AB023482 |
442 |
1e-121 |
splicing factor Prp8, Arabidopsis |
338 |
2e-92 |
|
BF256448 |
AB023482 |
SR |
SR |
multicatalytic endopeptidase complex, sequence removed |
SR |
SR |
|
BF256971 |
AP002071 |
434 |
1e-119 |
RNA-binding protein, Arabidopsis |
213 |
7e-55 |
|
BE603328 |
AB026295 |
SR |
SR |
unknown protein, sequence removed |
SR |
SR |
|
BE193581 |
AB026295 |
228 |
6e-57 |
MADS box protein, rice |
303 |
1e-81 |
|
|
AB023482 |
163 |
2e-37 |
homocitrate synthase, Thermus thermophilus |
166 |
2e-40 |
|
BE216383 |
AP000391 |
117 |
2e-23 |
hypothetical protein, rice |
248 |
8e-65 |
|
BF064968 |
AP000399; AP003487 |
321; 321 |
5e-85 5e-85 |
unknown protein, Arabidopsis |
125 |
3e-28 |
|
BF253546 |
AP002536 |
280 |
1e-72 |
serine / threonine phosphatase, Nicotiana
tabacum |
229 |
1e-69 |
|
BE194136 |
AP002536 |
147 |
1e-32 |
prolyl aminopeptidase-like protein, Arabidopsis |
198 |
2e-50 |
|
BF621935 |
AP002071; AP002854 |
248 248 |
5e-63
5e-63 |
unknown protein,
Arabidopsis |
100 |
9e-21 |
|
BF623965 |
AP002542; AP000559 |
281 664 |
3e-73 0.0 |
arginine decarboxylase, rice |
305 |
1e-82 |
|
BF625333 |
AP000399 |
404 |
1e-110 |
hexose carrier protein HEX6, rice |
258 |
2e-68 |
|
BF629359 |
AP002854 |
214 |
8e-53 |
seed imbibition, Sip1, barley |
243 |
1e-82 |
|
BF628496 |
AP002542; AP000559 |
353 353 |
1e-94 1e-94 |
Mg-protoporphyrin IX methyltransferase, rice |
266 |
1e-70 |
|
BF631507 |
AP000399 |
133 |
3e-28 |
similar serine/threonine protein kinase, rice |
233 |
2e-60 |
|
BF630362 |
AB023482 |
88 |
9e-15 |
kinase-like protein, Arabidopsis |
113 |
6e-25 |
|
BF257002 |
AP000399 |
90 |
2e-15 |
similar cytochrome c oxidase subunit Vb precursor, rice |
161 |
1e-50 |
|
BF267518 |
AP002854 |
305 |
2e-80 |
40S ribosomal protein S30, Arabidopsis |
124 |
4e-28 |
|
BF263245 |
AB023482 |
167 |
2e-38 |
gamma adaptin 1, rice |
386 |
1e-108 |
|
BF263968 |
AP000399 |
90 |
1e-15 |
no significant similarities |
|
|
|
BE215412 |
AP002542 |
276 |
2e-71 |
EPSP synthase, rice |
208 |
2e-53 |
|
BG414713 |
AP003632 |
162 |
6e-37 |
no good hits |
|
|
|
BG415252 |
AP000399 |
305 |
3e-80 |
hypothetical protein, rice |
376 |
1e-103 |
|
|
|
|
|
|
|
|
Table 1 continued
|
(Acc.#) |
(Acc. #) |
EST-PAC S-value |
EST-PAC
E-value |
Predicted Gene Function |
Function S -value |
E -value |
|
BF622807 |
AP003632 |
160 |
2e-36 |
nucleosome assembly protein, Arabidopsis |
218 |
4e-56 |
|
BF627428 |
AP003767 |
237 |
1e-59 |
proline-rich protein APG-isolog, Arabidopsis |
333 |
1e-90 |
|
BF629541 |
AP003526; AP003708 |
188 |
6e-45 |
no goods hits |
|
|
|
BG343171 |
AP003708 |
172 |
3e-40 |
homeodomain leucine zipper protein, rice |
164 |
9e-41 |
|
AW982580 |
AP003526 |
194 |
9e-47 |
kinesin-like protein, Arabidopsis |
309 |
2e-83 |
|
BG343499 |
AP003708 |
117 |
2e-23 |
serine/threonine protein kinase, Arabidopsis |
172 |
4e-42 |
|
BE601610 |
AP003526 |
162 |
3e-37 |
putative protein, Arabidopsis |
209 |
9e-54 |
|
BF257258 |
AP003708 |
190 |
9e-46 |
peptide methionine sulfoxide reductase, Arabidopsis |
271 |
3e-72 |
|
BF260645 |
AP003487 |
135 |
7e-29 |
carboxypeptidase, Arabidopsis |
258 |
4e-68 |
|
BF620281 |
AP003526 |
187 |
1e-44 |
hypothetical protein, Arabidopsis |
265 |
3e-70 |
|
BF261716 |
AP003767 |
233 |
2e-58 |
MAP kinase kinase, Arabidopsis |
171 |
3e-47 |
|
BF263248 |
AP000399 |
252 |
6e-64 |
arginine N-methyl transferase 1, Arabidopsis |
226 |
5e-72 |
|
BF263807 |
AP003767 |
210 |
1e-51 |
unknown protein, Arabidopsis |
169 |
2e-41 |
|
BF265658 |
AP003708 |
446 |
1e-122 |
root cap-specific protein, Maize |
328 |
5e-89 |
|
BE213851 |
AP003708 |
131 |
1e-27 |
hypothetical protein, Arabidopsis |
285 |
6e-76 |
|
BE214156 |
AP003487 |
82 |
1e-12 |
DNA-binding protein p24, Arabidopsis |
90 |
3e-17 |
|
BF065761 |
AP003708 |
218 |
7e-54 |
hypothetical protein, Arabidopsis |
273 |
1e-72 |
|
BF254888 |
AB026567c |
337 |
1e-89 |
beta 5 subunit of 20S proteasome, rice |
202 |
2e-51 |
|
BF623457 |
AP003454 |
146 |
3e-32 |
unknown protein, Arabidopsis |
109 |
3e-23 |
|
BF254110 |
AP003454 |
277 |
1e-71 |
putative protein, Arabidopsis |
135 |
8e-31 |
|
BE194388 |
AP002071; AP002854 |
149 149 |
4e-33 4e-33 |
beta-transducin-like protein, Arabidopsis |
348 |
3e-95 |
|
BE602073 |
AB026295 |
168 |
4e-39 |
unknown protein, rice |
199 |
1e-50 |
|
BG418108 |
AP002069 |
137 |
1e-31 |
hypothetical protein, rice |
243 |
8e-64 |
|
BF261612 |
AP002542 |
735 |
0.0 |
transketolase, rice |
402 |
1e-114 |
|
BF261621 |
AP002536 |
220 |
2e-54 |
serine/threonine protein phosphatase, alfalfa |
142 |
3e-33 |
|
BF266534 |
AP004239 |
485 |
1e-134 |
putative lipase, Arabidopsis |
226 |
1e-58 |
|
BE195608 |
AP000399 |
208 |
6e-51 |
early nodulin protein, rice |
134 |
2e-30 |
|
AW926067 |
AP000559; AP000391 |
SR |
SR |
receptor-like protein kinase, sequence removed |
SR |
SR |
|
AW926171 |
AP000559 |
SR |
SR |
Histone H3.2,minor, sequence removed |
SR |
SR |
|
SR =
sequence has been removed from the database. |
|
|
||||
|
c = accession # corresponds to a mRNA rice sequence
homologous to the barley EST. |
|
|||||
Table 2. Chromosome location(s) of barley EST loci that map away from the high-recombination region.
|
Chr-bin; locus |
(Acc.#) |
(Acc. #) |
EST-PAC S-value |
EST-PAC
E-value |
Predicted gene Function |
Function S-value |
Function E-value |
|
1 (7H) - 002 |
BE455209 |
AP000391 |
246 |
2e-62 |
similar to lipase |
303 |
1e-81 |
|
1 (7H) - 002 |
AW926881 |
AP000559 |
SR |
SR |
arginine decarboxylase |
SR |
SR |
|
1 (7H) - 002 |
AW925350 |
AP000559; AP002542 |
SR |
SR |
hypothetical protein |
SR |
SR |
|
1 (7H) - 002 |
BE195261 |
AP002542 |
? |
|
granule bound starch synthase |
196 |
8e-50 |
|
1 (7H) - 002 |
BF066009 |
AP000391 |
254 |
1e-64 |
hypothetical protein |
72 |
4e-26 |
|
1 (7H) - 002 |
BE060779 |
AP000559; AP002542 |
76 76 |
5e-11
5e-11 |
beta-I,3-glucanase |
227 |
8e-57 |
|
1 (7H) - 005 |
BF625282 |
AP003019 |
143 |
2e-31 |
ribosomal protein 60S- L39 |
109 |
1e-23 |
|
1 (7H) - 006 |
BF257721 |
AP000559 |
511 |
1e-142 |
Histone H3.2,minor |
266 |
1e-70 |
|
1 (7H) - 006 |
BE455049 |
AB023482 |
86 |
4e-14 |
no good hits |
|
|
|
1 (7H) - 007 |
BE060921 |
AP000559; AP002542 |
292; 292 |
2e-76; 2e-76 |
NAM-like protein |
258 |
3e-68 |
|
1 (7H) - 007 |
BF065540 |
AB023482 |
342 |
1e-91 |
hypothetical protein |
167 |
4e-41 |
|
1 (7H) - 007 |
AW983378 |
AP003487; AP003767 |
187; 187 |
2e-44; 2e-44 |
pectate lyase |
173 |
2e-42 |
|
1 (7H) - 007; A |
AW926171 |
AP000559 |
SR |
SR |
Histone H3.2,minor, sequence removed |
SR |
SR |
|
1 (7H) - 007; B |
BF622807 |
AP003632 |
160 |
2e-36 |
nucleosome assembly protein, Arabadopsis |
218 |
4e-56 |
|
1 (7H) - 008 |
BE215945 |
AP000399 |
121 |
5e-25 |
acyl-ACP thioesterase |
154 |
3e-37 |
|
1 (7H) - 010 |
BG344471 |
AP003708 |
164 |
1e-37 |
beta-1 subunit of 20S proteasome |
321 |
8e-89 |
|
1 (7H) - 012 |
BF259572 |
AP003019 |
473 |
1e-131 |
inorganic pyrophosphatase |
303 |
1e-81 |
|
1 (7H) - 012 |
BF626991 |
AP002071 |
SR |
SR |
hypothetical protein |
SR |
SR |
|
1 (7H) - 013 |
BG417554 |
AP003767 |
SR |
SR |
|
SR |
SR |
|
2 (2H) - 003 |
BG299346 |
AP003487 |
139 |
3e-30 |
sulfate transporter |
254 |
3e-67 |
|
2 (2H) - 005 |
BF616634 |
AP002542 |
278 |
7e-72 |
unknown protein |
311 |
5e-84 |
|
2 (2H) - 006 |
BE060723 |
AP002864 |
100 |
1e-18 |
Tubulin beta-3 chain |
320 |
1e-106 |
|
2 (2H) - 008 |
BG417957 |
AP003526 |
85 |
9e-14 |
no good hits |
|
|
|
2 (2H) - 008; B |
BE060893 |
AP002542 |
206 |
2e-50 |
40S ribosomal protein S20, rice |
218 |
2e-56 |
|
2 (2H) - 009 |
BF254704 |
AB026295 |
145 |
7e-32 |
HSPC133 protein |
155 |
6e-37 |
|
2 (2H) - 010 |
BF256092 |
AP000391 |
SR |
SR |
no significant similarities |
SR |
SR |
|
2 (2H) - 011 |
BF625997 |
AP002854 |
230 |
1e-57 |
alternative oxidase |
253 |
9e-74 |
|
2 (2H) - 013 |
BF624194 |
AB023482 |
178 |
3e-42 |
AP2 domain-containing protein |
133 |
8e-31 |
|
2 (2H) - 013 |
BF265753 |
AP003019 |
252 |
6e-64 |
H+ transporting ATPase |
338 |
2e-92 |
|
2 (2H) - 015 |
BG366491 |
AP003526 |
291 |
5e-76 |
putative protein |
173 |
5e-43 |
|
2 (2H) - 015 |
BF256699 |
AP003767 |
223 |
2e-55 |
no good hits |
|
|
|
3 (3H) - 006 |
BF617975 |
AP000399 |
119 |
3e-24 |
arginine N-methyl transferase |
137 |
4e-32 |
|
3 (3H) - 008; A 4 (4H) - 001; B 5 (1H) - 010; C |
BE214588 |
AP002864 |
117 |
1e-23 |
tubulin beta-3 chain |
407 |
1e-113 |
Table 2 continued
|
Chr-bin; locus |
(Acc.#) |
(Acc. #) |
EST-PAC S-value |
EST-PAC
E-value |
Predicted gene Function |
Function S-value |
Function E-value |
|
3 (3H) - 011 |
BF257999 |
AP000399 |
SR |
SR |
|
SR |
SR |
|
4 (4H) - 002 |
BF258878 |
AP000559;
AP002542; AP002071 |
SR |
SR |
unknown protein |
SR |
SR |
|
4 (4H) - 002; A 4 (4H) - 010; B |
BF258346 |
AP002854 |
490 |
1e-135 |
hypothetical protein |
101 |
9e-21 |
|
5 (1H) - 006; A 6 (6H) - 006; B |
BF065140 |
AP000399 |
78 |
1e-11 |
unknown protein |
224 |
5e-58 |
|
5 (1H) - 007 |
BG369940 |
AP003526; AP003487 |
SR |
SR |
no good hits |
SR |
SR |
|
5 (1H) - 008 |
BG367156 |
AP002069 |
294 |
4e-77 |
ubiquinol cytochrome c reductase |
144 |
5e-34 |
|
5 (1H) - 008 |
BG414283 |
AP002542 |
162 |
4e-37 |
beta-tonoplast intrinsic protein |
201 |
3e-51 |
|
5 (1H) - 011; A 7 (5H) - 006; B |
AW983097 |
AB023482 |
82 |
1e-12 |
no good hits |
|
|
|
5 (1H) - 013 |
BE060078 |
AP002542 |
86 |
7e-22 |
unknown protein |
74 |
1e-12 |
|
6 (6H) - 005; B |
BF261716 |
AP003767 |
233 |
2e-58 |
MAP kinase kinase, Arabadopsis |
171 |
3e-47 |
|
6 (6H) - 006 |
BF253463 |
AP002864 |
117 |
1e-23 |
Tubulin beta-2 chain |
244 |
8e-64 |
|
6 (6H) - 006 |
BG299297 |
AP002542 |
241 |
8e-61 |
granule-bound starch synthase precursor |
350 |
8e-96 |
|
6 (6H) - 014 |
BG343190 |
AP002854 |
188 |
6e-45 |
TAT-binding protein |
359 |
3e-98 |
|
6 (6H) - 014; A 3 (3H) - 015; B |
BG344873 |
AP002854 |
180 |
1e-42 |
TAT-binding protein |
324 |
4e-88 |
|
7 (5H) - 002; A 7 (5H) - 004; B |
BE602168 |
AP003526 |
112 |
5e-22 |
enolase |
441 |
1e-123 |
|
7 (5H) - 006 |
BF258338 |
AP000399 |
593 |
1e-167 |
hypothetical protein |
249 |
2e-72 |
|
7 (5H) - 009 |
BE195592 |
AP002069 |
SR |
SR |
ethylene-forming-enzyme-like dioxygenase |
SR |
SR |
SR = sequence has been removed from the database.