
High-density mapping of a gene-rich region
present on homoeologous group 5L of
wheat and barley
M. Maroof Shah and Kulvinder S. Gill
University of Nebraska, P. O. Box 830915, Lincoln, NE 68583
Email - kgill@unl.edu
Tel.- 402-472-1534, Fax- 402-472-7904
Wheat and barley belong to the grass family Poaceae, which also includes rice,
oat and maize. Deletion lines-based physical maps in wheat (Gill et al., 1996a,b) and
translocation breakpoint-based physical maps in barley (Kunzel et al., 2000) revealed that
most genes are present in physically small gene-rich regions. These gene-rich regions
are interspersed by large chromosomal blocks mainly containing repetitive sequences
(Barakat et al., 1997). One such gene-rich region is present in the middle of wheat
homoeologous group 5 long arm chromosome. The region contains genes controlling an
array of important agronomic traits. Among these is Ph1, the principal gene regulating
chromosome pairing, gene for barley short rachilla hair (mSrh), high lysine (Lys1),
aleurain (Ale), reaction to Erysiphe graminis f.sp.hordei (Mlj), Chlorina seedling (f6), and
hybrid chlorosis (Chl). The region also contains genes controlling maturity, plant height,
grain yield and its component traits.
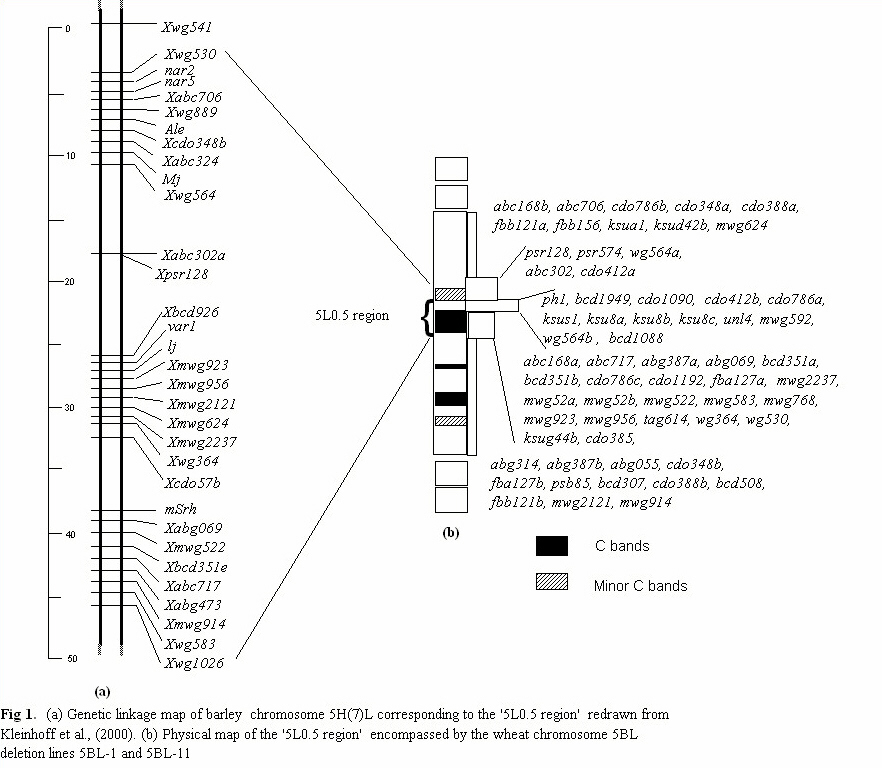
In wheat, the region is best localized on chromosome 5BL where it is flanked by
the breakpoints of deletions 5BL-1 and 5BL-11 between fraction length (FL) 0.55 and
0.59 (Gill et al., 1996a). The corresponding region in barley is present between fraction
length (FL) 0.68 and 0.71 of chromosome 5HL (Kunzel, 2000., Shah et al., 1999). This
region will be referred to as the '5L0.5 region'. The Ph1locus is further localized to a
smaller chromosomal region bracketed by 5BL-1 on the proximal end and the distal
breakpoint of the interstitial deletion of mutant ph1b on the distal side (Gill et al., 1993).
This region will be called "Ph1 gene region". The objectives in the present study were to
enrich '5L0.5 region' with markers by comparative mapping and to study its structure
between wheat and barley.
We identified 75 DNA markers for the region by comparing 36 different Poaceae
maps. In the first phase of comparative mapping, RFLP probes, previously mapped in
the region (Gill, 1993; Gill, 1996a], were used as anchor markers. Newly identified
probes for the region were used as anchors to perform a second round of comparative
mapping. Fifty-seven out of the total 75 markers were tested by the gel blot DNA
hybridization analysis on the blots containing HindIII and DraI or EcoRI digested DNA
of the three nullisomic-tetrasomic lines for wheat homoeologous group 5 chromosomes
[NT5A(5B), NT5B(5A), NT5D(5A)], Ph1 mutants [ph1b, Ph1dup], and 5BL deletion
lines [5BL-1, and 5BL-11]. The Ph1 mutant lines were used to assign the DNA fragment
bands to the 'ph1 gene region'. The gel blot DNA analysis was performed as previously
described (Gill, 1993). Probes detecting 5B DNA fragment bands that were absent in the
deletion line 5BL-1 but present in 5BL-11, were mapped to the '5L0.5 region'. Probes
detecting a 5BL chromosome specific fragment that was not present in the ph1b mutant
and was also missing in the deletion line 5BL-1 but present in the 5BL-11 were the 'Ph1
gene region' markers.
A total of 61 marker loci were detected by physical mapping of 47 RFLP probes
on the wheat chromosome 5B (Fig.1b). Of those, 39 marker loci mapped in the '5L0.5
region'. Twelve of these 39 loci mapped in the 'ph1 gene region'.
The physical map around the '5L0.5 region' was compared with the barley
physical map (Kunzel, 2000) and the genetic linkage map developed in the Steptoe X
Morex (S/M) population (Kleinhofs., 2000) (Fig.1a). Thirteen marker loci were common
between the barley genetic linkage map of chromosome 5H and the physical map
generated for '5L0.5 region' of wheat. The marker order between the two maps appeared
to be similar except for the probe wg530. The probe mapped in the '5L0.5 region' on the
physical map. On the barley genetic map, however, it is flanked by the markers (abc706
and wg541) present in the proximal 20% of the arm. The probe wg530 detects four
bands in wheat and nine bands in barley. The mapping discrepancy may be due to the
presence of another locus. Recombination in the region was very high (Fig.1 a, b).
Genetic length of the '5L0.5 region' in barley was about 45cM out of the total map
length of 145 cM for chromosome 5HL (Kleinhofs, 2000).
These results demonstrate the power of comparative mapping in Triticeae for
characterizing physically smaller regions of the genomes. The comparative mapping
strategy was very effective as 39 of the 61 probes mapped in the '5L0.5 region'. Among
the '5L0.5 region' loci, 12 mapped in the 'Ph1 gene region' making it more amenable to
target the Ph1 locus. Currently we are in the process of screening the barley BAC library
using the '5L0.5 region' markers in an attempt to construct a contig map of the region.
References
Barakat A., N. Carels, and G. Bernardi. 1997. The distribution of genes in the genomes of Graminae. Proc Natl Acad Sci USA 94: 6857-6861
Gill, K. S., B. S. Gill, T. R. Endo, E. V. Boyko. 1996a. Identification and high-density mapping of gene-rich regions in chromosome group 5 of wheat. Genetics 143:
1001-1012
Gill, K. S., B. S. Gill, T. R. Endo, and Y. Mukai. 1993. Fine physical mapping of Ph1, a chromosome pairing regulator gene in polyploid wheat. Genetics, 134:
1231-1236.
Gill, K. S., B. S. Gill, T. R. Endo, and T. Taylor. 1996b. Identification and high-density mapping of gene-rich regions in chromosome group 1 of wheat. Genetics 144:
1883-1891.
Kleinhofs, A. 2000. Barley maps . http://barleygenomics.wsu.edu/Macdraw/7-150.jpg
Kunzel, G,, L. Korzum, and A.Meister. 2000. Cytologically integrated physical restriction fragement length polymorphism maps for the barley genome based on
translocation breakpoints. Genetics 154: 397-412
Shah, M. M., and K. S. Gill. 2000. Marker enrichment and fine mapping of the barley 5H(7) chromosomal region homologous to the Ph1 gene of wheat. Barley NewsLetter Vol.43;http://grain.jouy.inra.fr/ggpages/BarleyNewsletter/43/shah.html

