Deric Schmierer1, Nejdet Kandemir2, David Kudrna1, Darrell Wesenberg3,
Steve Ullrich1, Andris Kleinhofs1,4
1Department of Crops and Soil Sciences, Washington State University, Pullman, WA, 99164, USA;
2Gaziosmanpasa Universitesi, Ziraat Fakultesi, Tarla Bitkileri Bolumu, 60110, Tokat, Turkey;
3National Small Grains Germplasm Research Facility, USDA-ARS, Aberdeen, ID, 83210, USA;
4School of Molecular Biosciences, Washington State University, Pullman, WA, 99164, USA
The American malting industry two-row malting barley standard cv. Harrington tends to yield significantly lower than cv. Baronesse, especially in dryland growing conditions. Baronesse, a high-yielding two-row feed barley, was originally developed in Europe, but has adapted very well to the growing conditions in the PNW. Baronesse accounted for approximately 70% of the total barley acreage grown in Washington in 1998 and over 74% in 1999 covering nearly 370,000 acres in both years. This cultivar is the leading feed barley grown in the state. Harrington was developed in Canada and accounted for almost 7% of the total barely acreage in the state for both 1998 and 1999. It is the leading malting barley grown in Washington, planted on about 35,000 acres in both years. Similar numbers have been reported in Oregon, Idaho, and Montana, with Harrington and Baronesse being the top malting and feed barleys, respectively. Our goal is to introduce the good yield and adaptability characteristics of Baronesse into new lines while maintaining the good malting characteristics of Harrington.
Baronesse yield QTL were mapped to the long arm of barley chromosome 2- (2HL) and chromosome 3- (3HL) (T. Blake, personal commun.). Using a molecular marker-assisted (MAS) backcross breeding scheme, a population of near isogenic lines (NILs) were produced from a Harrington x Baronesse cross. Two experiments were conducted involving two and three backcross schemes, respectively. After each backcross and selfing operation, progeny were selected based on their genotypes using RFLP markers in the chromosome 2HL and 3HL regions.
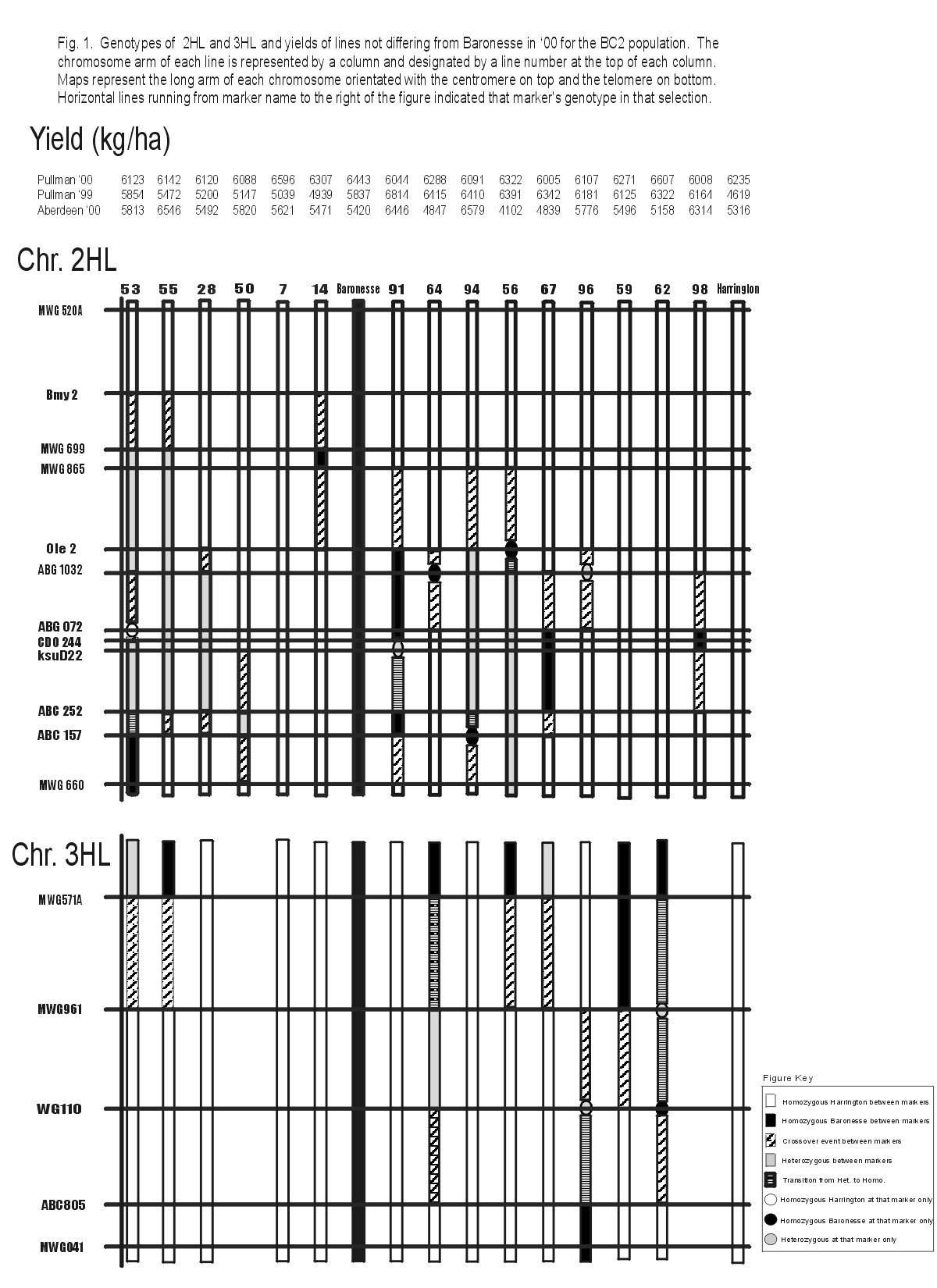
In 1999, 110 BC2 lines were planted in replicated yield trials at Spillman Farm in Pullman, WA. 53 lines produced yields similar to Baronesse. Of these 53 lines, 12 yielded significantly higher than Harrington. In summer 2000, 28 high yielding lines from the 1999 BC2 trials were planted in replicated yield trials at Pullman and Aberdeen, ID. Of the 28 BC2 lines tested, 16 yielded similar to Baronesse. Six lines yielded higher than Harrington, though differences were not significant (Fig. 1). Based on these two years of BC2 yield data and genotypes of these lines, two regions on chromosome 2HL and one region on chromosome 3HL are suggested as containing potential high yield QLTs. These regions are between Bmy2 - Ole2 and ABG1032 - ABC252 on 2HL and between MWG571A - WG110 on 3HL (Fig. 1). One interesting note is that line 7 does not contain any Baronesse alleles in these target regions, yet yielded significantly higher than Harrington at Pullman in both 1999 and 2000. This suggests that there are other Baronesse yield QTLs that have not been identified.
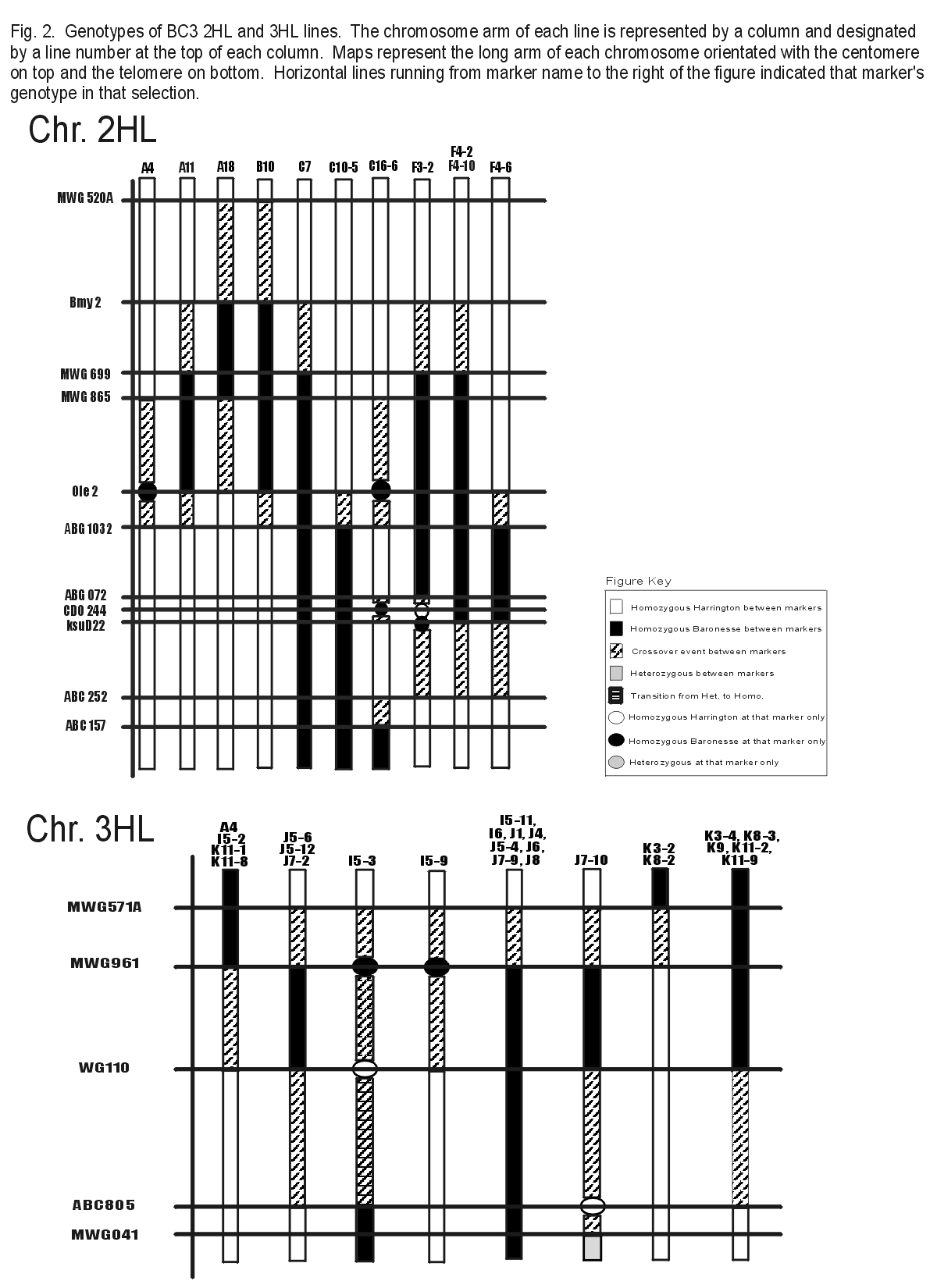
In summer 2000, 84 lines from the BC3 population were grown in replicated yield trials at Pullman and Aberdeen. 48 of these 84 lines yielded similar to Baronesse. 22 of these 48 lines also yielded significantly higher than Harrington (Table 1). Genotyping and yield data of the BC3 lines narrowed the 2HL and 3HL chromosome regions that confer yield QTL. The 3HL region is now estimated to be between MWG571A - MWG961. The proximal region of 2HL appears to reside between Bmy2 - MWG699 and the distal region between ABG072 - ksuD22 (Fig. 2). Several lines that seem to contain the 2HL distal region yielded significantly lower than Baronesse (data not shown). This suggests that the proximal region has a larger effect. Another explanation is that other regions of the genome contained within these lines have a detrimental effect on yield.
Yield data from Aberdeen was not as informative as Pullman data for all populations. Harrington and Baronesse performed in an opposite manner than expected under irrigation, with Harrington yielding higher, or not different, than Baronesse. Although this effect was observed, several lines yielded near the top at Pullman and Aberdeen in 2000 (Table 2). These lines may be worthy of further investigation in low rainfall, irrigated areas.
Preliminary yield data from Pullman in 1999 and 2000 suggests that we have accomplished our goal of increasing the yield of Harrington by transferring Baronesse yield QTL. Although regions of chromosomes 2HL and 3HL have been targeted as potentially carrying yield QTL, there must be other Baronesse QTL in regions of the genome yet to be detected. Malting analysis is pending.
| Table 1. BC3 lines that did not yield significantly different than Baronesse at Pullman in 2000. Italicized lines yielded significantly higher than Harrington. |
| ID | Avg. kg | Avg. kg | Chromosome genotyped | Baronesse fragment of genotyped
chromosome (crossovers occurred outside of indicated markers)
| |||
| Nursery2 | |||||||
| A18-5 | 7422 | 672.7 | 2H | Bmy2-MWG865 | |||
| A11-1 | 7342 | 686.8 | 2H | MWG699-Ole2 | |||
| BARONESSE | 7211 | 711.7 | |||||
| A18-1 | 7193 | 668.8 | 2H | Bmy2-MWG865 | |||
| C16-6 | 7114 | 676.5 | 2H | Ole2; CDO244; ABC157 | |||
| A11-3 | 6857 | 680.8 | 2H | MWG699-Ole2 | |||
| C7-2 | 6852 | 689.0 | 2H | MWG699-ABC157 | |||
| A18-2 | 6822 | 665.0 | 2H | Bmy2-MWG865 | |||
| B10-6 | 6754 | 678.7 | 2H | Bmy2-Ole2 | |||
| A11-2 | 6727 | 678.7 | 2H | MWG699-Ole2 | |||
| C7-3 | 6587 | 690.7 | 2H | MWG699-ABC157 | |||
| B10-1 | 6579 | 677.0 | 2H | Bmy2-Ole2 | |||
| A4-19 | 6564 | 679.1 | 2H, 3H | 2H: Ole2; 3H: MWG571A-MWG961 | |||
| HARRINGTON | 5715 | 679.5 | |||||
| Nursery 3 | |||||||
| J4-3 | 7392 | 672.2 | 3H | MWG961-MWG041 | |||
| J6-5 | 7314 | 667.1 | 3H | MWG961-MWG041 | |||
| J6-6 | 7259 | 670.5 | 3H | MWG961-MWG041 | |||
| J5-4 | 7083 | 669.2 | 3H | MWG961-MWG041 | |||
| J5-6 | 7076 | 675.7 | 3H | MWG961-WG110 | |||
| J4-1 | 7073 | 669.7 | 3H | MWG961-MWG041 | |||
| BARONESSE | 7063 | 700.6 | |||||
| J6-4 | 6966 | 674.8 | 3H | MWG961-MWG041 | |||
| I5-9 | 6961 | 680.0 | 3H | MWG961 | |||
| J1-3 | 6942 | 669.2 | 3H | MWG961-MWG041 | |||
| I6-1 | 6757 | 673.1 | 3H | MWG961-MWG041 | |||
| J4-2 | 6752 | 669.7 | 3H | MWG961-MWG041 | |||
| J1-2 | 6710 | 663.2 | 3H | MWG961-MWG041 | |||
| I5-2 | 6701 | 673.5 | 3H | MWG571A-MWG961 | |||
| HARRINGTON | 6694 | 675.3 | |||||
| F4-2 | 6686 | 677.0 | 2H | MWG699-ksuD22 | |||
| I5-11 | 6628 | 667.5 | 3H | MWG961-MWG041 | |||
| F4-6 | 6624 | 677.4 | 2H | ABG1032-ksuD22 | |||
| I5-3 | 6602 | 670.1 | 3H | MWG961; ABC805-MWG041 | |||
| F4-10 | 6587 | 677.8 | 2H | MWG699-ksuD22 | |||
| J5-12 | 6520 | 669.7 | 3H | MWG961-WG110 | |||
| I6-3 | 6486 | 663.2 | 3H | MWG961-MWG041 | |||
| F3-2 | 6457 | 670.1 | 2H | MWG699-ABG072; ksuD22 | |||
| Nursery 4 | |||||||
| K11-1 | 7399 | 665.8 | 3H | MWG571A-MWG961 | |||
| K9-2 | 7100 | 675.7 | 3H | MWG571A-WG110 | |||
| BARONESSE | 7092 | 694.1 | |||||
| J6-8 | 7067 | 666.7 | 3H | MWG961-MWG041 | |||
| K3-2 | 6957 | 679.5 | 3H | MWG571A | |||
| K9-1 | 6866 | 682.1 | 3H | MWG571A-WG110 | |||
| J7-9 | 6857 | 670.5 | 3H | MWG961-MWG041 | |||
| J7-2 | 6721 | 671.8 | 3H | MWG961-WG110 | |||
| J7-10 | 6721 | 668.0 | 3H | MWG961-WG110; MWG041* | |||
| J8-3 | 6596 | 676.5 | 3H | MWG961-MWG041 | |||
| K3-4 | 6568 | 677.8 | 3H | MWG571A-WG110 | |||
| K8-3 | 6556 | 681.2 | 3H | MWG571A-WG110 | |||
| K11-2 | 6530 | 676.1 | 3H | MWG571A-WG110 | |||
| J8-2 | 6502 | 674.0 | 3H | MWG961-MWG041 | |||
| K8-2 | 6467 | 672.7 | 3H | MWG571A | |||
| K11-8 | 6452 | 678.2 | 3H | MWG571A-MWG961 | |||
| HARRINGTON | 6012 | 674.4 |
| * MWG041 is heterozygous in line J7-10 |
| Table 2. BC3 lines that yielded near the top at both Pullman and Aberdeen in '00. |
| ID | Pullman '00 Avg. kg ha-1 |
Aberdeen '00 Avg. kg ha-1 |
Chromosome genotyped |
Baronesse fragment of genotyped
chromosome (crossovers occurred outside of indicated markers) |
| A11-3 | 6857 | 6859.0 | 2H | MWG699-Ole2 |
| J6-4 | 6966 | 6673.0 | 3H | MWG961-MWG041 |
| A11-1 | 7342 | 6558.0 | 2H | MWG699-Ole2 |
| K9-2 | 7100 | 6531.0 | 3H | MWG571A-WG110 |
| C16-6 | 7114 | 6523.0 | 2H | Ole2; CDO244; ABC157 |
| A18-1 | 7193 | 6513.0 | 2H | Bmy2-MWG865 |
| A18-5 | 7422 | 6483.0 | 2H | Bmy2-MWG865 |
| A18-2 | 6822 | 6475.0 | 2H | Bmy2-MWG865 |
| C7-2 | 6852 | 6352.0 | 2H | MWG699-ABC157 |
| K3-2 | 6957 | 6164.0 | 3H | MWG571A |
| K9-1 | 6866 | 6055.0 | 3H | MWG571A-WG110 |
| J6-5 | 7314 | 5909.0 | 3H | MWG961-MWG041 |
| J5-6 | 7076 | 5861.0 | 3H | MWG961-WG110 |
| J6-8 | 7067 | 5840.0 | 3H | MWG961-MWG041 |
| J6-6 | 7259 | 5805.0 | 3H | MWG961-MWG041 |
| J5-4 | 7083 | 5671.0 | 3H | MWG961-MWG041 |
| J4-3 | 7392 | 5667.0 | 3H | MWG961-MWG041 |
| J7-9 | 6857 | 5648.0 | 3H | MWG961-MWG041 |
| J4-1 | 7073 | 5239.0 | 3H | MWG961-MWG041 |