
Efficient cloning of resistance gene analogs from barley
Robert Brueggeman1, Arnis Druka1, David Kudrna1 and Andris Kleinhofs1,2
1 Depts. of Crop and Soil Sciences, Washington State University, Pullman, WA, 99164-6420
2 Depts. of Molecular Biosciences, Washington State University, Pullman, WA, 99164-6420
In recent years disease resistance genes (R-genes) have been cloned and characterized
from a variety of plants including both mono- and dicotyledon species. This growing list of R-genes has revealed that a majority belongs to the NBS-LRR class of genes which are
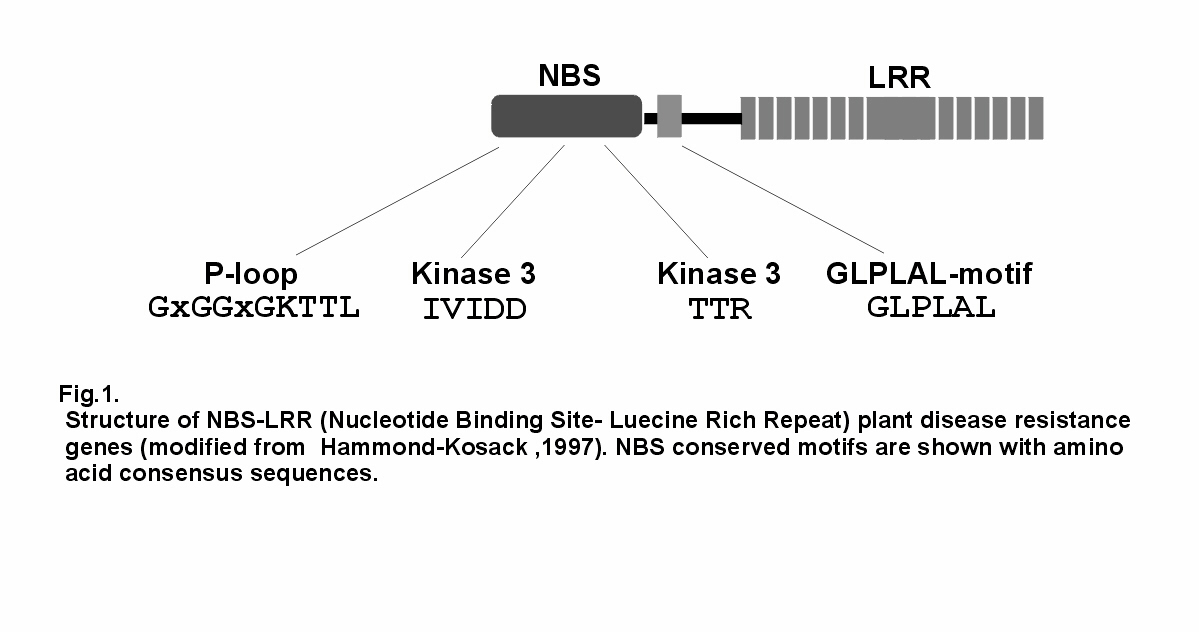
characterized by a nucleotide-binding site (NBS) near the N-terminus and a leucine-rich repeat
(LRR) region near the C-terminus (Fig 1). An increasing number of NBS-LRR encoding
sequences are also being identified through PCR amplification with degenerate primers, EST
sequencing, BAC end sequencing and genome sequencing projects. The Arabidopsis Genome
Initiative reported that the Arabidopsis genome is estimated to contain approximately 200 genes
encoding NBS-LRR related motifs. This represents close to 1% of all Arabidopsis genes (The
Arabidopsis Genome Initiative, 2000). In rice it has been predicted that NBS-LRR encoding
sequences may be even more prevalent than in Arabidopsis (Meyers et al., 1999). The wide
distribution of these sequences in the plant kingdom and their prevalence in the Arabidopsis and
rice genomes indicates that they are ancient, diverse and common in plants. Their abundance in
plant genomes and the belief that NBS-LRR encoding genes confer pathogen recognition
triggering defense mechanisms suggests that they play a significant role in plant disease
resistance. Identifying these resistance gene analog (RGA) sequences in barley and mapping
them could prove to be important in the identification of actual R-genes as well as areas of the
genome were R-genes may reside. These cloned RGAs will be a valuable resource as markers
for molecular breeding programs as well as excellent probes for the identification of RGAs or R-genes in other cereal crops.
Overall sequence homology among R-genes of the NBS-LRR class is low, but the NBS
contains several sequence motifs, P-loop, kinase2, kinase3 and GLPLAL, that are highly
conserved among R-genes even in distantly related plants (Hammond-Kosack and Jones, 1997).
Synthetic oligonucleotides designed from these highly conserved motifs have been used to
amplify and clone RGAs from plant genomic DNA. However, in large genome species this
procedure also generates large numbers of DNA fragments with no apparent homology to R-genes. In order to improve barley RGA cloning efficiency, we performed three methods: 1)
Hybridization of NBS derived oligonucleotides to arrayed BAC and cDNA libraries, 2) PCR
amplification of RGAs from BAC DNA pools, and 3) PCR amplification of RGAs from cDNA
libraries. All methods yielded confirmed RGA clones, but experiments involving hybridization
yielded many artifacts or non-RGA genes containing NBS-like motifs. Hybridization to either
the BAC or cDNA libraries recovered RGA containing clones with only a 3% efficiency.
Experiments involving PCR amplification from BAC DNA pools were more successful giving
44% efficiency, but the best results were obtained by PCR amplification from cDNA libraries
which gave 52% efficiency. All methods were more efficient than PCR from genomic DNA,
which had an efficiency of about 1%. All confirmed RGAs were genetically mapped and BAC
clones isolated. Subcloning weakly hybridizing bands identified in BAC clones discovered
additional new RGAs.
Two previously undescribed RGAs, ABG331 and RSB001, were identified by labeling
NBS derived oligonucleotides and utilizing them as hybridization probes on the BAC library (Yu
et al., 2000). BACs identified were further screened by PCR and the resulting fragments of the
expected size were cloned and sequenced. From this hybridization procedure 68 clones have
been sequenced and two were RGAs, giving a RGA cloning efficiency of about 3%. RSB001
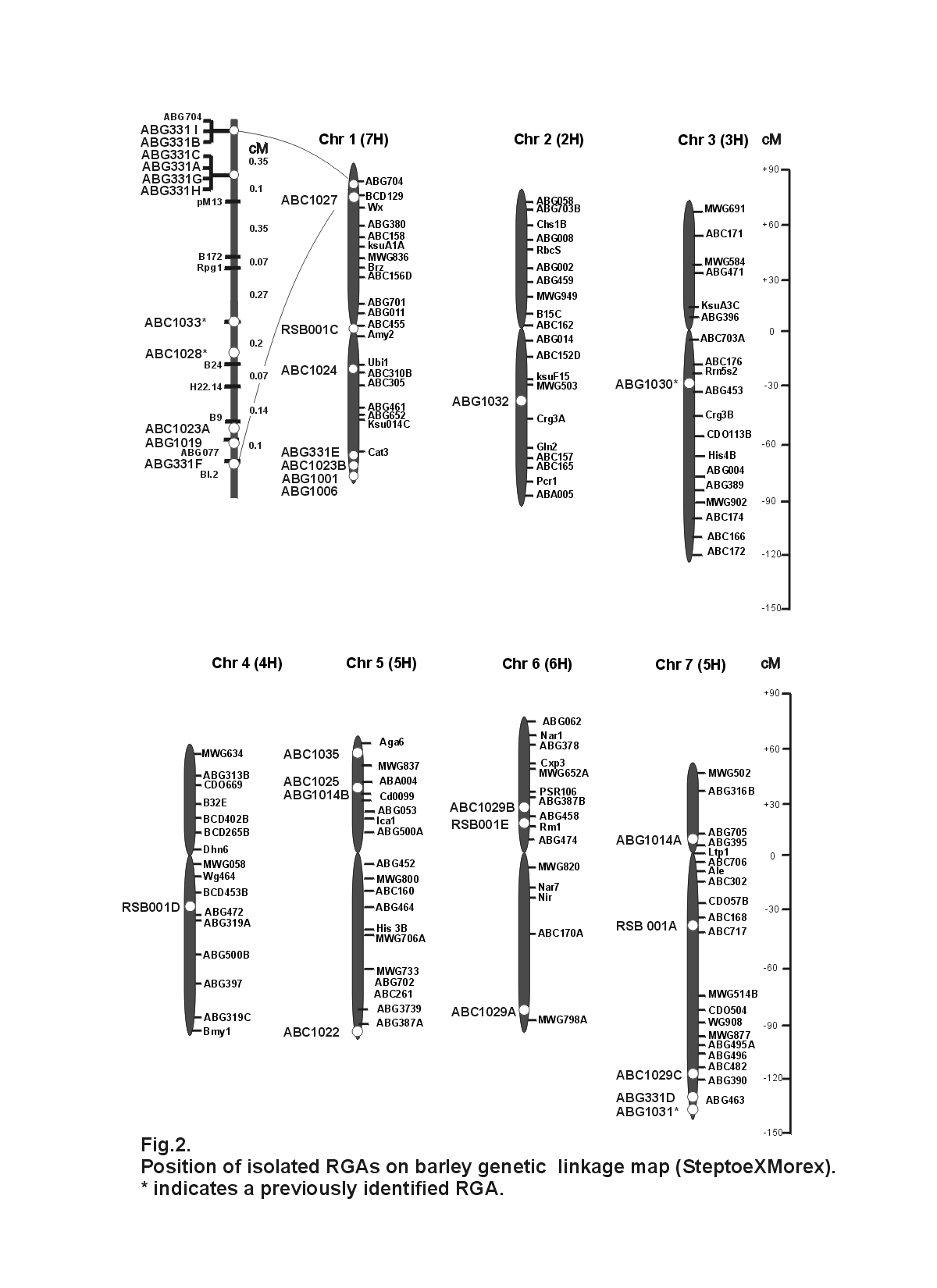
identified a gene family that mapped to loci on chromosomes 1,2,4,6 and 7(Fig 2) and possibly
other yet undetermined loci. ABG331 mapped to eight loci on chromosome 1 and one locus on
chromosome 7(Fig 2). One RGA was identified by NBS oligo hybridization to an arrayed cDNA
library. Thirty-three cDNAs identified were sequenced and one was a RGA, giving an efficiency
of 3%. This RGA, ABC1024, had been previously identified by PCR amplification using a
cDNA library as template.
Five new RGAs, ABG1001, ABG1006, ABG1019, ABG1014 and ABG1032, were
identified by PCR amplification with the NBS derived oligonucleotides using BAC clone pools
as template. From this amplification procedure 68 clones of the expected size have been
sequenced and 28 were RGAs, giving an efficiency of about 44%. ABG1014 mapped to loci on
chromosomes 5 and 7(Fig 2). ABG1001, ABG1006, ABG1019 and ABG1032 each mapped to a
single locus (Fig 2).
Six new RGAs, ABC1022, ABC1023, ABC1024, ABC1025, ABC1027, ABC1029, and
ABC1035 were identified by PCR amplification from cDNA libraries. From this procedure 61
clones were sequenced and 32 were RGAs, giving an efficiency of 52%. ABC1023 mapped to
two loci on chromosome 1 (Fig 2). ABC1029 mapped to two loci on chromosome 6 and one
locus on chromosome 7(Fig 2). ABC1022, ABC1024, ABC1027 and ABC1035 each mapped to
a single locus (Fig 2). ABC 1025 is the cDNA equivalent of ABG1014. BAC clones
corresponding to the mapped loci were identified and the RGA-related fragments subcloned and
analyzed by sequence comparison with known NBS-LRR genes. These methods allowed us to
identify new RGA clones, for example ABG1034, which had not been mapped to date. These
RGAs could have utility in the isolation of new RGAs and perhaps actual disease resistance
genes.
References
The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering
plant Arabidopsis thaliana. Nature, 408, 796-815.
B.C. Meyers, A.W. Dickerman, R.W. Michelmore, S. Sivaramakrishnan, B.W. Sobral and N.D.
Young (1999) Plant disease resistance genes encode members of an ancient and diverse
protein family within the nucleotide binding superfamily. The Plant Journal, 20, 317-332.
K.E. Hammond-Kosack and J.D.G. Jones (1997) Plant disease resistance genes. Annu. Rev.
Plant Physiol. Plant Mol. Biol. 48, 575-607.
Y. Yu, J.P. Tomkins, R. Waugh, D.A. Frisch, D. Kudrna, A. Kleinhofs, R.S Brueggeman, G.J.
Muehlbauer, R.P. Wise, R.A. Wing (2000) A bacterial artificial chromosome library for
barley (Hordeum vulgare L.) and identification of clones containing putative resistance
genes. Theor. Appl. Genet., 101, 1093-1099.


