H. Salvo G. and J. W. Snape
John Innes Centre, Norwich Research Park
Colney, Norwich, NR4 7UH, UK
Abstract
A genetic map of diploid Hordeum bulbosum cross was generated (Jaffe et. al. 2000; Salvo et. al. in preparation) and is now being used for QTL analysis of traits in H. bulbosum. Here, preliminary results of QTL analysis for crossability between H. bulbosum and H. vulgare are presented. Homoeologous regions within barley and wheat appeared to have a significant association with different crossability traits on chromosomes 3Hb, 5Hb and 7Hb.
Introduction
Hordeum bulbosum is crossable with barley and thus has two major uses in cultivated barley improvement. First, it is used for high frequency haploid production in breeding programs to speed up the breeding cycle, and, secondly for alien gene introgression. Greater knowledge of the genome organization and the control of specific traits would facilitate its efficient uses for both.
To date only one H. bulbosum linkage map has been developed (Jaffe et. al. 2000; Salvo et. al.), but this can now be applied to analyse the crossability with H. vulgare. There is a major problem in QTL mapping in outcrossing species such as H. bulbosum which concerns the fact that two to four alleles may be involved in the segregation within the outcrossing family and this may vary between markers. In the BC1-type of segregation only one of the parents can provide information with respect to linked QTLs, whereas in the other types the meiosis of both parents are informative. In the case of markers with an F2 segregation type it is unknown for the heterozygous progeny which allele was derived from which parent. Thereby the ideal markers for QTL mapping are those segregating for 3 or 4 alleles, essentially because all parental gametes can be recognized unambiguously for all progeny genotypes (Maliepaard and Van Ooijen, 1994)
This paper presents preliminary results of a first experiment to examine QTL analysis for crossability in H. bulbosum. A series of traits concerned with crossability between H. vulgare and H. bulbosum were measured in a extensive crossing programme.
Materials and methods
By forced cross-pollination a recombinant population was obtained from a cross of clones PB1 and PB11 of diploid Hordeum bulbosum (2n=2x=14). A range of cDNA, and genomic DNA clones from barley, wheat, rice and oats, available in the Cereals Research Department, John Innes Centre, were used as probes. Most probes used had been mapped in barley (Laurie et al. 1995), and other species, and anchored on the maps of wheat, barley and rice. The notation for the segregation of markers follows Maliepaard et al. (1997). The different alleles of a marker locus were characterized with different letters, such as a, b, c, d, including '0' for a null-allele.
The Hordeum bulbosum vernalization, crossing, embryo rescue and in vitro culture techniques were carried out as described by Simpson and Snape (1981), Pickering (1982) and Furusho et al. (1990). However, in this experiment a controlled environment was used, with the aim of increasing the relative effect of genetic variation among the recombinant clones within the population by decreasing the environmental variation. Pollinated spikes were detached from each stem and put in water containing nutrient solution under controlled environmental conditions at 22±2°C day /17±2°C night; 16 hour day length and relative humidity 75±5%. Under these conditions, after 17 days, embryo rescue was made. For in vitro culture, the FWG regeneration media was used. This is a modified Murashige and Skoog (1962) medium with lower NH4NO3 and high Mg SO4; supplemented with 20 gl-1 maltose, 4 mgl-1 thiamine, 1gl-1 myo-inositol and 7.5 gl-1 glutamine.
The experimental design was a completely randomized design with four replications, where each replication was represented by one spike with 20 flowers. Crosses were randomized over plant and days.
For QTL analysis, crossability percentages of each trait were transformed by arcsin. To look for the location of QTL for crossability, the transformed data were analyzed using single factor analyses of variance for each marker locus over recombinant clones to detect significant associations (P< 0.05) between a marker locus and the traits.
Results and Discussion
A total of 2,960 barley florets were pollinated with H. bulbosum pollen from different recombinant clones. Genetic variation among the male parents was noted for several traits associated with crossability: seeds set per flower (S/F), embryos produced per seed (E/S) and germination frequency, plants per embryo (P/E) and callus formation frequency per embryo (C/E). In relation to the frequency distributions for the different traits, S/F presented a normal distribution, and all the other traits gave transgressive segregation, which might indicate that dispersed genes between the parents are controlling these traits.
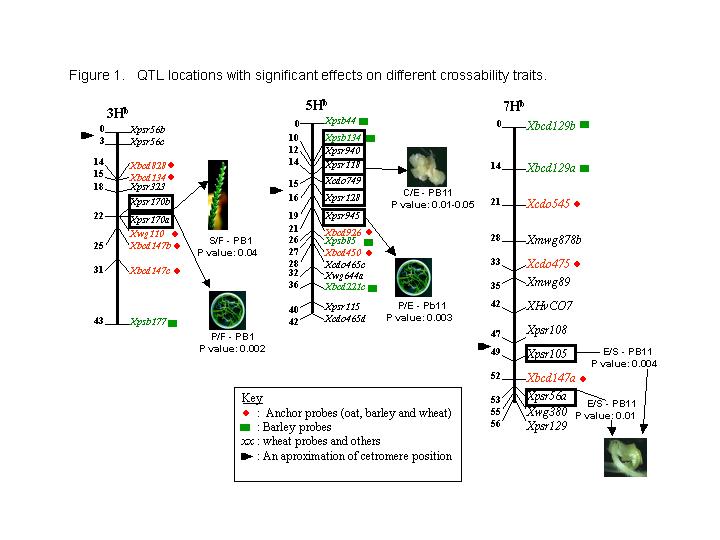
In this first screen for QTL, some markers linked to genomic regions of H. bulbosum were shown to have significant effects, see Figure 1. It is interesting to note that in chromosome 5Hb some molecular markers gave a significant association for interspecific crossability. Those markers are very close to Xpsr128, a marker already mapped as being close to a QTL for interspecific crosability between H. bulbosum (2n=4x=28) and wheat recombinant lines in chromosome 5BL of wheat (Zhang et al. 1998). On the same genomic region, the marker Xcdo749 has also been reported with a significant association to crossability between wheat and barley (Taketa et al. 1998). The same author found a QTL for crossability in the long arm of barley chromosome 3H, a region where the barley cDNA anchor probe Xbcd828 was located in the middle of the confidence interval of the QTL. In H. bulbosum, this locus has been mapped in the same homoeologous genomic region 3HbL, close to the locus Xpsr170 (6 cM), which in this first screen for QTL has been found highly significant effect on crossability among plant per flowers (0.002). Another marker, Xpsr105, presented a highly significant association (0.004) for embryos per seed; markers mapped close to centromere on chromosome 7Hb. This could indicate that this chromosome in H. bulbosum also has some genes controlling interspecific crossability with H. vulgare.
To summarise, it is important to point out that QTL analysis for crossability is one of the important and immediate uses for our H. bulbosum map. In this first screening for QTLs some genomic region on H. bulbosum chromosomes appeared to have a significant effect on crossability with H. vulgare. Some of them could be homoeologous regions within the Triticeae, and it may be possible that there are orthologous genes controlling these specific traits. These results are being confirmed and will give an important insight into the efficient use of H. bulbosum in haploid production and the introgression from this wild species into barley.
Acknowledgements
This work is a part of a PhD programme, which is supported by INIA, Chile (National Institute of Agriculture Research) and ORS awards in the U. K.
References
Chen Z., Devey M., Tuleen N. A., Hart G. E. 1994. Use of recombinant substitution lines in the construction of RFLP-based genetics maps of chromosomes 6A and 6B of tetraploid wheat (Triticum turgidum L.). Theor Appl Genet. 89:703-712.
Furusho, M., Hamachi, Y. and Yoshida, T. 1990. Varietal difference in crossability between Japanese two-rowed barley and Hordeum bulbosum L. Japan. J.Breed. 40:411-417.
Laurie, D. A., Pratchentt, N. Bezant, J. Snape, J. W. 1995. RFLP mapping of 13 genes controlling flowering time in a winter x spring barley (Hordeum vulgare L.) cross. Genome, 38:575-585.
Maliepaard C., Janse J., Van Ooijen J. W. 1997. Linkage analysis in full-sib family of an outbreeding plant species: Overview and consequences for applications. Genet. Res., Camb. 70: 237-250.
Maliepaard C., Van Ooijen J. W. 1994. QTL mapping in a full-sib family of an outcrossing species In: Biometrics in plant breeding: Applications of molecular markers. J.W. van Ooijen and J. Jensen ed. Proc. of the 9th Meeting of the EUCARPIA Section Biometrics in Plant Breeding 6-8 July. Wageningen, the Netherlands. pp. 140-146.
Murashige, T., and Skoog, F. 1962. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15: 473-479.
Pickering, R. A. 1982. The effect of pollination bag type on seed quality and size in Hordeum Inter- and intraspecific hybridation. Euphytica 31: 439-449.
Simpson, E. and Snape, J. W. 1981. The use of doubled haploids in winter barley programme. In: Barley Genetics IV. Edinburgh, University Press. Edinburgh. 716-720.
Taketa S., Takahashi H., Takeda K. 1998. Genetic variation in barley crossability with wheat and its quantitative trait loci analysis. Euphytica, 103:187-193.
Van Deynze A. E., Sorrells M.E., Park W. D., Ayres N. M., Fu H., Cartinhour S. W., Paul E., McCouch S. R., 1998. Anchor probes for comparative mapping of grass genera. Theor Appl Genet. 97:356-369.
Zhang W., Zheng Y., Foote T., Doldersum J., Dunford R., Fish L., Wang Y., Moore G., Snape J. 1998. Mapping key genes on chromosome 5B of wheat. In: John Innes Centre & Sainsbury Lab. Annual Report 1997/98. p. 12.