

REPORTS
OF THE COORDINATORS
Overall coordinator’s
report
Udda Lundqvist
Svalöf Weibull AB
SE-268 81 Svalöv, Sweden
Since
the latest overall coordinator’s report in Barley Genetics Newsletter Volume
29, no important changes of the coordinators have been reported. I do hope that
most of you are willing to continue with this work and provide us with all new
information you can find in the literature and get from other researchers about
the chromosomes, linkage groups and collections over the year. In this
connection I want to stress again as I did earlier that all information is of
great importance for the barley community. Many younger researchers are dealing
with molecular genetics, different mapping programs are going on world-wide and
many new techniques are available and used. Especially the coordinator for
”Integrating Barley Molecular and Morphological/Physiological Maps” has a very
important and heavy task and responsiblity following all the information in the
literature and updating the maps. He is also doing important research in his
own laboratory and has provided us with useful maps in the last issues of
Barley Genetics Newsletter. Unhappily, very little research is going on with
many of the morphological and chromosomal collection groups, therefore the
reports are very short and mostly a reminder that they still exist.
Also
in this issue the reports of the seven barley chromosomes are arranged
according to the resolution made at the Seventh International Barley Genetics
Symposium in Saskatoon, Canada, in 1996. This year no revised and new
descriptions of different morphological and physiological traits have been
compiled. Therefore, no new current lists of BGS descriptions by BGS number and
by locus symbol in alphabetic order are published in this issue. Those
published in the last volumes of BGN are still valid and up-to-date.
All
information regarding morphological marker stock is available electronically at
the following addresses:
1.
http://www.ars-grin.gov/ars/PacWest/Aberdeen
2. http://wheat.pw.usda.gov/ggpages/bgn
In
about half a year the VIIIth International Barley Genetics Symposium will be
organized in Adelaide, South Australia. I hope that many of you will be able to
participate in the meetings. I would like to encourage the coordinators and
their colleagues already today to provide me with ideas, aspects, items or
topics which should be brought up during the conference. One important item has
to be discussed if the coordination system should continue as it exists today
or if it should be organized in some other form.
Chromosome 1H (5): Jens Jensen, Plant Biology and
Biogeochemistry Department, Risø National Laboratory, P.O. Box 301, DK-4000
Roskilde, Denmark. FAX: +45 46 77 4122; e-mail: <jens.jensen@risoe.dk>
Chromosome 2H (2): Jerry. D. Franckowiak, Department of
Plant Sciences, North Dakota State University, P.O.Box 5051, Fargo, ND
58105-5051, USA. FAX: +1 701 231 8474; e-mail: <jfrancko@badlands.nodak.edu>
Chromosome 3H (3): Takeo Konishi, 294 Okada, Mabi-cho,
Kibi-gun, Okayama 710-1311, Japan. FAX: +81 866 98 4334; e-mail: <konishit@okym.enjoy.ne.jp>.
Chromosome 4H (4): Brian P. Forster, Cell and Molecular
Genetics Department, Scottish Crop Research Institute, Invergowrie, Dundee, DD2
5DA, United Kingdom. FAX: +44 1382 562426. e-mail: <bforst@scri.sari.ac.uk>
Chromosome 5H (7): George Fedak, Eastern Cereal and Oilseed
Research Centre, Agriculture and Agri-Food Canada, ECORC, Ottawa, ON, Canada
K1A 0C6, FAX: +1 613 759 6559; e-mail: <fedakga@em.agr.ca>
Chromosome 6H (6): Duane Falk, Department of Crop Science,
University of Guelph, Guelph, ON, Canada, N1G 2W1. FAX: +1 519 763 8933;
e-mail: <dfalk@crop.uoguelph.ca>
Chromosome 7H (1): Lynn Dahleen, USDA-ARS, State University
Station, P.O. Box 5677, Fargo, ND 58105, USA. FAX: + 1 701 239 1369; e-mail:
<DAHLEENL@fargo.ars.usda.gov>
Integration of molecular
and morphological marker maps:
Andy Kleinhofs, Department of Crop and Soil Sciences, Washington State
University, Pullman, WA 99164-6420, USA. FAX: +1 509 335 8674; e-mail: <andyk@wsu.edu>
Barley Genetics Stock
Center: An Hang and
Darell M. Wesenberg, USDA-ARS, National Small Grains Germplasm Research
Facility, P.O.Box 307, Aberdeen, ID 83210, USA. FAX: +1 208 397 4165; e-mail:
<anhang@uidaho.edu>
Trisomic and aneuploid
stocks: An Hang,
USDA-ARS, National Small Grains Germplasm Research Facility, P.O.Box 307,
Aberdeen, ID 83210, USA. FAX: +1 208 397 4165; e-mail: <anhang@uidaho.edu>
Translocations and balanced
tertiary trisomics:
Gottfried Künzel, Institute of Plant Genetics and Crop Plant Research,
Corrensstrasse 3, DE-06466 Gatersleben, Germany. FAX: +49 39482 5137; e-mail:
<kuenzel@ipk-gatersleben.de>
Desynaptic genes: Gottfried Künzel, Institute of Plant
Genetics and Crop Plant Research, Corrensstrasse 3, DE-06466 Gatersleben,
Germany. FAX: +49 39482 5137; e-mail: <kuenzel@ipk-gatersleben.de>
List of Barley Coordinators
(continued)
Autotetraploids: Wolfgang Friedt, Institute of Crop
Science and Plant Breeding, Justus-Liebig-University, Ludwigstrasse 23, DE-35390
Giessen, Germany. FAX: +49 641 9937429; e-mail: <wolfgang.friedt@agrar.uni-giessen.de>
Disease and pest resistance
genes: Brian Steffenson,
Department of Plant Pathology, North Dakota State University, P.O. Box 5012,
Fargo, ND 58105-5012, USA. FAX: +1 701 231 7851; e-mail: <bsteffen@badlands.nodak.edu>
Eceriferum genes: Udda Lundqvist, Svalöf Weibull AB,
SE-268 81 Svalöv, Sweden. FAX:.+46 418 667109; e-mail: <udda@ngb.se>
Chloroplast genes: Diter von Wettstein, Department of Crop
and Soil Sciences, Genetics and Cell Biology, Washington State University,
Pullman, WA 99164-6420, USA. FAX: +1 509 335 8674; e-mail: <diter@wsu.edu>
Genetic male sterile genes: Mario C. Therrien, Agriculture and
Agrifood Canada, P.O. Box 1000A, R.R. #3, Brandon, MB, Canada R7A 5Y3, FAX: +1
204 728 3858; e-mail: <mtherrien@em.agr.ca>
Inversions: Bengt-Olle Bengtsson, Institute of
Genetics, University of Lund, Sölvegatan 29, SE-223 62 Lund, Sweden. FAX: +46
46 147874;
e-mail: <bengt-olle.bengtsson@gen.lu.se>
Anthocyanin genes: Barbro Jende-Strid, Department of
Physiology, Carlsberg Research Laboratory, Gamle Carlsberg Vej 10, DK-2500
Copenhagen-Valby, Denmark. FAX: +45 33 274764; e-mail: <bjs@crc.dk>
Ear morphology genes: Udda Lundqvist, Svalöf Weibull AB,
SE-268 81 Svalöv, Sweden. FAX: +46 418 667109; e-mail: <udda@ngb.se> and
Arne
Hagberg, Department of Plant Breeding Research, The Swedish University of
Agricultural Sciences, SE-268 31 Svalöv, Sweden. FAX: +46 418 667081; e-mail:
-.
Semi-dwarf genes: Jerry D. Franckowiak, Department of
Plant Sciences, North Dakota State University, P.O. Box 5051, Fargo, ND
58105-5051, USA. FAX: +1 702 231 8474; e-mail: <jfrancko@badlands.nodak.edu>
Early maturity genes: Udda Lundqvist, Svalöf Weibull AB,
SE-268 81 Svalöv, Sweden. FAX: +46 418 667109; e-mail: <udda@ngb.se>
Chromosome duplications: Arne Hagberg, Department of Plant
Breeding Research, The Swedish University of Agricultural Sciences, SE-268 31
Svalöv, Sweden. FAX: +46 418 667081; e-mail: -.
List of Barley Coordinators
(continued)
Monoclonal antibodies: Steven E. Ullrich, Department of Crop
and Soil Sciences, Washington State University, Pullman, WA 99164-6420, USA.
FAX: +1 509 335 8674; e-mail: <ullrich@wsu.edu>
Biochemical mutants -
Including lysine, hordein and nitrate reductase: Andy Kleinhofs, Department of Crop and
Soil Sciences, Washington State University, Pullman, WA 99164-6420, USA. FAX:
+1 509 335 8674; e-mail: <andyk@wsu.edu>
Barley-wheat genetic
stocks: A.K.M.R. Islam,
Department of Plant Science, Waite Agricultural Research Institute, The
University of Adelaide, Glen Osmond, S.A. 5064, Australia. FAX: +61 8 8303
7109; e-mail: <rislam@waite.adelaide.edu.au>
Coordinator's Report:
Barley Chromosome 5 (1H)
Jens Jensen
Plant Biology and Biogeochemistry Department,
PBK-301,
Risø National Laboratory
DK-4000 Roskilde, Denmar
A
high-resolution map at the Mla locus
using AFLP markers was developed to isolate YAC clones harbouring the Mla locus (Schwarz et al., 1999).
A fine resolution map of molecular markers
between the loci Hor1 and Hor2 was used to detect three families
of resistance-gene homologues of the nucleotide-binding site /leucine-rich
repeat type at the Mla locus region
(Wei et al., 1999).
The most telomeric region of the barley
chromosome 5 (1H) had the markers XBA1 and
ABA305, designated Tel5L and Tel5S, respectively, indicating the long and short arm of the
chromosome. Furthermore, the marker ABR337
was found to be located at the centromere (Kilian et al., 1998).
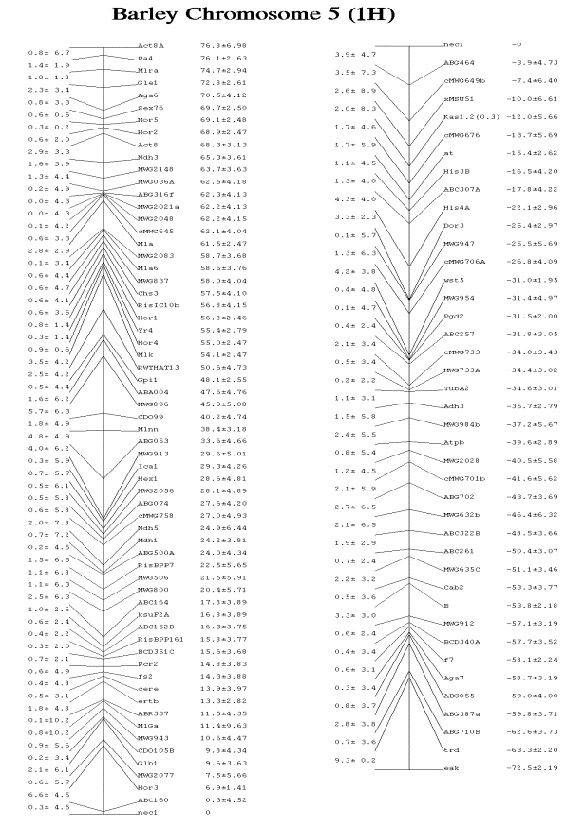
No revision of the chromosome 5 (1H)
linkage map was necessary, therefore the unchanged map is shown in figure 1 as
it was published in the last issue of BGN 29.
References
Kilian,
A., D. Kudrna and A. Kleinhofs. 1999. Genetic and molecular characterization of
barley telomeres. Genome 42:412-419.
Schwarz,
G., W. Michalek, V. Mohler, G. Wenzel and A. Jahoor. 1999. Chromosome landing
at the Mla locus in barley (Hordeum
vulgare L.) by means of high-resolution mapping with AFLP markers. Theor.
Appl. Genet. 98:521-530.
Wei, F.,
K. Gobelman-Werner, SM. Morroll, J. Kurth, L. Mao, R. Wing, D. Leister, P.
Schulze-Lefert and RP. Wise. 1999. The Mla
(powdery mildew) resistance cluster is associated with the three NBS-LRR
gene families and suppressed recombination within a 240-kb DNA interval on chromosome
5S (1HS) of barley. Genetics 153:1929-1948.
Fig.1. (following page) The barley
chromosome 5 linkage map is calculated based on the linkage information
reported in the former issues of BGN and no changes have been made since last
years report. The map positions are given in centimorgans (cM). The distances
between neighbouring loci are given to the left, the loci names and positions
are placed to the right.
Coordinator's
report: Chromosome 2H (2)
J.D. Franckowiak
Department of Plant Sciences, North
Dakota State University
Fargo, ND 58105, U.S.A.
Larson et al., 1998 mapped a low phytic acid (lpa1) mutant to chromosome 2HL of barley and reported that the lpa1 locus was not related to any of the
seven orthologous myo-inositol 1‑phosphate synthase (MIPS) candidate
genes in maize. But, they suggested based on RFLP mapping data that the Ipa1 region of barley is homoeologous to
the Ipa1 region of maize chromosome
1S. The lpa1.1 gene does not alter
total phosphorus (P) content of barley kernels, but it causes a 50% reduction
in phytic acid P and a corresponding increase in inorganic P (Raboy and Cook
1999). The P in phytic acid is unavailable to humans and non‑ruminants
and produces waste P that can runoff as water pollution. Unlike the lpa2 mutants, plants with the lpa1.1 gene appear to have normal growth
and productivity (Raboy and Cook, 1999).
Börner et al., 1999 mapped the position of two recessive dwarfing mutants
in chromosome 2H. The gibberellic acid (GA) insensitive dwarf, gai (Hv287),
which co-segregated with RFLP markers MWG2058 and MWG2287, is close to the
centromere in chromosome 2HS. The GA sensitive dwarf, gal (Hv288), which co-segregated with RFLP markers MWG581 and
MWG882A, is in chromosome 2HL, about 22 cM distal from the six-rowed spike 1 (vrs1)
locus and about 55 cM distal from the gai
locus. Komatsuda et al., 1999
published a high resolution map for the region around the six-rowed spike 1 (vrs1) locus in chromosome 2HL.
Pickering et al., 2000 located in chromosome 2HL of line 38P18 a DNA segment
from Hordeum bulbosum that contains a
gene for resistance to Puccinia hordei.
Line 38P18, derived from a cross between Emir and H. bulbosum, produces a fleck reaction to all leaf rust isolates
tested. Previously Pickering et al.,
1998 reported that line 81882 has in chromosome 2HS a DNA segment from H. bulbosum that confers leaf rust and
powdery mildew resistance. Line 81882, derived from a Vada cross, produces a
low infection type when inoculated with leaf rust isolates. Both lines show
reduced plant vigor under field conditions. The locus name Rph17 and allele symbol Rph17.af
are suggested for the leaf rust resistance gene in line 81882. The locus name Rph18 and allele symbol Rph18.ag are suggested for the leaf rust
resistance gene in line 38P18.
Wheat-barley addition lines were used in
two studies to associate barley genes with chromosome 2H. Hansson et al., 1998 determined the chromosome
arm locations of six enzymes for chlorophyll and haem synthesis including
placement of the magnesium chelatase subunit Xantha-F in chromosome 2HS. Nomura
et al., 1999 found that the presence
of Hordatines A and B, strong antifungal components, in barley seedlings is
associated with chromosome 2HS.
Borem et
al. 1999 studied starch granules in the barley endosperm and found that a
region of chromosome 2 contains QTLs affecting the overall mean granule volume,
the proportion of type A (large) granules, the mean volume of type A granules,
the mean maximum diameter of type A granules, and the mean F-shape of type B
(small) granules.
Data on photoperiod responses in barley
were published in two papers. Karsai et
al., 1997 studied heading date in the Dicktoo/Morex mapping population and
reported that QTLs for earliness are associated with the early maturity 1 (Eam1 or Ppd-H1) locus in chromosome 2HS and with the spring growth habit 2
(Sgh2 or Sh2) locus in chromosome 5HL. Alleles at the Eam1 (Ppd-H1) locus were
found to vary in their response to photoperiod duration. Stracke and Börner,
1998 presented segregation data for photoperiod responses under short-day
conditions of F2 and F3 plants from a Atsel/Betzes cross.
Atsel and Betzes have contrasting alleles at the Eam1 locus in chromosome 2HS and at the eam7 locus in chromosome 6HS. Their results indicated that the
recessive eam7.g allele from Atsel
causes photoperiod insensitivity when the dominant allele at the Eam1 locus, also from Atsel, is present.
Zhu et
al., 1999 collected Fusarium head blight (FHB) response data from several
environments for doubled-haploid lines from a Gobernadora/CMB643 cross. Data
were gather also for several morphological traits. They found that the Vrs1.t (deficiens) allele in chromosome
2HL was associated QTLs for resistance to FHB and more seeds per spike. The Int-c.a allele at the intermedium-c
locus in chromosome 4HS was associated with susceptibility to FHB, large
lateral spikelets, and shorter plants. Both the Vrs1.t and the Int-c.a
alleles were derived from the cultivar Shyri via CMB643. The parental lines for
this cross were developed by the CIMMYT/ICARDA barley breeding program in
Mexico.
Kleinhofs, 1997 using bulk segregate
analyses associated several morphological markers of barley with specific
molecular markers. Later, Kleinhofs et al.,
1998 presented the BIN method for subdividing the barley molecular marker maps
into segments, BINs of about 10 cM each. Information on the assignment of
markers to BINs is based on the Steptoe/Morex molecular marker map and is
available at http://barleygenomics.wsu.edu/
as an EXCEL spreadsheet download. File data and the results of Kleinhofs, 1997
were used to add several new markers to the morphological marker map for
chromosome 2H (Figure 1). Figure 1 also summarizes a few comparisons between
BIN assignments for several loci and their relative positions on the
morphological marker map. Although direct comparisons of the two maps are not
possible because the morphological marker map is based data accumulated from
many studies, the estimated position of two only two morphological markers does
not fit a linear gene order. The elongated outer glume (eog) locus is near the centromere, but it is associated with BIN 2‑004.
The morphological marker map places the triple awned lemma (trp) locus far beyond the last BIN of
chromosome 2HL.
References
Borem,
A., D.E. Mather, D.C. Rasmusson, R.G. Fulcher and P.M. Hayes. 1999. Mapping
quantitative trait loci for starch granule traits in barley. J. Cereal Sci. 29
(2):153-160.
Börner,
A., V. Korzun, S. Malyshev and V. Ivandic. 1999. Molecular mapping of two
dwarfing genes differing in their GA response on chromosome 2H of barley.
Theor. Appl. Genet. 99:670-675.
Hansson,
M., S.P. Gough, C.G. Kannangara and D. von Wettstein. 1998. Chromosomal
locations of six barley genes encoding enzymes of chlorophyll and heme
biosynthesis and the sequence of ferrochelatase gene identify two regulatory
genes. Plant Phys. Biochem. Paris 36(8):545-554.
Karsai,
I., K. Mezaros, P.M. Hayes and Z. Bedo. 1997. Effects of loci on chromosomes 2
(2H) and 7 (5H) on development patterns in barley (Hordeum vulgare L.) under different photoperiod regimes. Theor.
Appl. Genet. 94:612-618.
Kleinhofs, A. 1997.
Integrating barley RFLP and classical maps. BGN 27:105-112.
Kleinhofs,
A., D. Kudrna and D. Matthews. 1998. Integrating barley molecular and
morphological /physiological marker maps. BGN 28:89-91.
Komatsuda,
T., W. Li, F. Takaiwa and S. Oka. 1999. High resolution map around the vrs1 locus controlling two- and
six-rowed spike in barley, Hordeum
vulgare. Genome 42:248-253.
Larson,
S.R., K.A. Young, A. Cook, T.K. Blake and V. Raboy. 1998. Linkage mapping of
two mutations that reduce phytic acid content of barley grain. Theor. Appl.
Genet. 97:141‑146.
Larson,
S.R. and V. Raboy. 1999. Linkage mapping of maize and barley myo-inositol 1-phosphate
synthase DNA sequences corresponding with a low phytic acid mutation. Theor.
Appl. Genet. 99:27-36.
Nomura,
T., M. Sue, R. Horikoshi, S. Tebayashi, A. Ishihara, T.R. Endo and H. Iwamura.
1999. Occurrence of hordatines, the barley antifungal compounds, in a
wheat-barley chromosome addition line. Genes Genet. Systems 74:99-103.
Pickering,
R.A., S. Malyshev, G. Künzel, P.A. Johnston, V. Korzun, M. Menke and I.
Schubert. 2000. Locating introgressions of Hordeum
bulbosum chromatin within the H.
vulgare genome. Theor. Appl. Genet. 100:27-31.
Pickering,
R.A., B.J. Steffenson, A.M. Hill and I. Borovkova. 1998. Association of leaf
rust and powdery mildew resistance in a recombinant derived from a Hordeum vulgare x H. bulbosum hybrid. Plant Breed. 117:83-84.
Raboy,
V. and A. Cook. 1999. An update on ARS barley low phytic acid research. BGN 30:
http://wheat.pw.usda.gov/ggpages/bgn/29/a29‑08.html
Stracke,
S. and A. Börner. 1998. Molecular mapping of the photoperiod gene ea7 in barley. Theor. Appl. Genet. 97:797-800.
Zhu, H., L. Gilchrist, P. Hayes, A. Kleinhofs,
D. Kudrna, Z. Liu, L. Prom, B. Steffenson, T. Toojinda and H. Vivar. 1999. Does
function follow form? Principal QTLs for Fusarium head blight (FHB) resistance
are coincident with QTLs for inflorescence traits and plant height in a double
haploid population of barley. Theor. Appl.. Genet. 99:1221-1232.
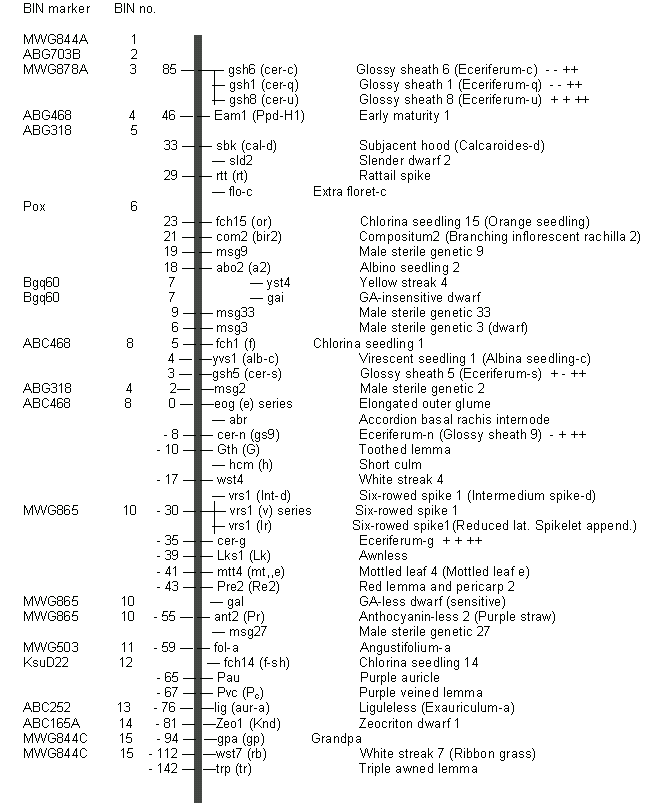
Figure 1. Estimated positions for morphological marker loci and corresponding BIN numbers in chromosome 2H of barley.
Coordinator's Report:
Chromosome 3H (3)
T. Konishi
294 Okada, Mabi-cho, Kibi-gun, Okayama
710-1311, Japan
Kilian
et al., 1999 mapped 33 different loci
generated from telomere-associated sequences using RFLP and PCR techniques, and
identified the most telomeric regions of 10 of the 14 barley chromosome arms. Tel3S (ABA307B) was located as a marker
to the map’s most terminal region of chromosome 3HS, whereas three markers,
generated by PCR using a single primer based on the sequence of the barley
telomere repeats, were mapped internally in chromosome 3HL. Cloning and
sequencing of PCR products from the 3HL interstitial location revealed homology
to the HvRT family of tandem repeats isolated from barley.
Dávila
et al., 1999 localized 35 random
amplified microsatellite polymorphism (RAMP) markers onto the Steptoe/Morex
RFLP map. RAMP markers were distributed over all the seven barley chromosomes,
and several RAMPs were mapped in large gaps of the RFLP linkage map, improving
the coverage of certain chromosome regions. Seven of RAMPs covered chromosomes3
(3H) as well as chromosomes 5 (1H) and 6 (6H) as a large number, and some of
the markers in these chromosomes were observed in small clusters around the
centromere.
Kleinhofs,
1999 made a great progress in the integration of the barley molecular maps with
the morphological and physiological markers using the Oregon World Barley
Dominant x Recessive doubled haploid (DH) population, and mapped more than 60
markers in the seven barley chromosomes with reasonable linkage estimates. Nine
of the markers, mo7a, alm, msg5,
uzu, wst6, wst1, als, sdw1
and Pub, were located in the
molecular map of chromosome 3H, ranging from the short arm to the long arm of
the chromosome. However, the
arrangement of the markers was somewhat different from the order in chromosome
3H illustrated by Franckowiak, 1996, especially the wst1 locus was tightly linked with the uzu locus (Takahashi and Moriya, 1969; Tsuchiya and Jensen, 1973).
Graner
et al., 1999 conducted the molecular
mapping of the rym5 locus encoding
resistance to barley mild mosaic virus (BaMMV) and two strains of barley yellow
mosaic virus (BaYMV-1 and BaYMV-2), using 391 DH lines derived from eight
crosses between a series of rym5 W122
lines and susceptible cultivars. The marker analysis revealed that rym5 could be placed into an interval
flanked by markers MWG838 and MWG10 near the distal end of chromosome 3HL, and
mapped 0.8% distal from MWG838 and 1.3% proximal from MWG10. This map position
is thought to be almost the same or extremely close to the rym4 locus controlling resistance to BaMMV, since rym4 is located 1.2 cM distal from
MWG838 and 1.2 cM proximal from MWG10 (Graner and Bauer, 1993). In fact, all
the BaMMV-resistant DH lines were resistant to BaYMV, while the
BaMMV-susceptible lines also exhibited susceptibility to BaYMV. No
recombination between BaMMV- and BaYMV-resistant DH lines suggests that rym4 and rym5 are tightly linked or allelic. The latter possibility might be
supported by the fact that F1 plants between ‘Franka’ (rym4) and W122/37.1 (rym5) were resistant to BaMMV in
Germany. However, it might be worthwhile commenting that ‘Franka’ was resistant
to the Natajima strain of BaMMV in Japan, whereas ‘Misato Golden’ (rym5) was susceptible (Konishi and
Kaiser, 1999). ‘Misato Golden’ and W122 lines contain the rym5 gene derived from the same parent, Mokusekko 3.
Raman
and Read, 1999 developed the efficient marker assisted selection for resistance
to barley yellow dwarf virus (BYDV). The resistance gene Ryd2 in chromosome 3HL is closely linked with the probe BCD828. As
RFLP analysis is expensive, laborious and involves radioisotopes, PCR based
assays were practiced using leaf tissue and sap as templates. The results were
in good quality as isolated ‘purified’ DNA, indicating that direct leaf
tissue/leaf sap could be used successfully as templates for marker assisted
selection for the resistance.
Boron
toxicity is an important problem limiting production in the low-rainfall
regions of southern Australia, West Asia and North Africa. Genetic variation
for boron toxicity tolerance in barley has been recognized, but the mode of
inheritance and the location of genes controlling tolerance were not known
previously. Jefferies et al., 1999
conducted interval regression-mapping of QTL’s for boron tolerance, using 150
DH lines from a cross between a boron toxicity tolerant Algerian landrace,
Sahara 3771, and the intolerant Australian cultivar, Clipper. The analysis
which revealed four regions in chromosomes 2H, 3H, 4H and 6H were associated
with boron tolerance. The region in the short arm of chromosome 3H was strongly
associated with root-length response to high boron concentration, together with
the region on the long arm of chromosome 4H. RFLP markers in the regions of
chromosomes 3HS and 4HL were xAWBMA15
and xWG114, respectively.
Dehydrins
comprising an immunologically distinct protein family are associated with
tolerance to drought and low temperature. Choi et al., 1999 identified 11 unique Dhn genes in the cv Dicktoo, and mapped the Dhn genes in the chromosomes 3H, 4H, 5H and 6H by PCR with
wheat-barley addition lines. The Dhn10
and Dhn11 genes were located in
chromosome 3H, both of them were newly detected in barley and their
counterparts are unknown in other plants.
References
Choi, D.W., B. Zhu and
T.J. Close. 1999. The barley (Hordeum
vulgare L.) dehydrin multigene family: sequences, allele types, chromosome
assignments, and expression characteristics of 11 Dhn genes of cv Dicktoo. Theor. Appl. Genet. 98:1234-1247.
Davila, J.A., Y. Loarce
and E. Ferrer. 1999. Molecular characterization and genetic mapping of random
amplified microsatellite polymorphism in barley. Theor. Appl. Genet.
98:265-273.
Franckowiak, J.D. 1996.
Revised linkage maps for morphological markers in barley, Hordeum vulgare. BGN 26:9-21.
Graner, A. and E. Bauer.
1993. RFLP mapping of the ym4 virus
resistance gene in barley. Theor. Appl. Genet. 86:689-693.
Graner,
A., S. Streng, A. Kellermann, A. Schiemann, E. Bauer, R. Waugh, B. Pellio and
F. Ordon. 1999. Molecular mapping and genetic fine-structure of the rym5 locus encoding resistance to different strains of the Barley
yellow Mosaic Virus Complex. Theo. Appl. Genet. 98:285-290.
Jefferies,
S.P., A.R. Barr, A. Karakousis, J.M. Kretschmer, S. Manning, K.J. Chalmers,
J.C. Nelson, A.K.M.R. Islam and P. Langridge. 1999. Mapping of chromosome
regions conferring boron toxicity tolerance in barley (Hordeum vulgare L.). Theor. Appl. Genet. 98:1293-1303.
Konishi,
T. and R. Kaiser. 1999. Reaction of barley accessions to BaMMV and BaYMV in
Japan, compared with data in Germany. BGN 30 (this volume).
Kilian
A., D. Kudrna and A. Kleinhofs. 1999. Genetic and molecular characterization of
barley chromosome telomeres. Genome 42:412-419.
Kleinhofs,
A. 1999. Coordinator’s report: Integrating barley molecular and
morphological/physiological marker maps. BGN 29:
Raman,
H. and B.J. Read. 1999. Efficient marker assisted selection for resistance to
barley yellow dwarf virus using leaf tissue and sap as templates in barley. BGN
29:
Takahashi,
R. and I. Moriya. 1969. Inheritance and linkage studies in barley. IV. Linkages
of four variegated mutants. Ber. Ohara Inst. landw. Biol., Okayama Univ.
15:35-46.
Tsuchiya,
T. and D.A. Jensen. 1973. Further results on the allelic relationship between wst and wst3. BGN 3:69-70.
Coordinator's report: Chromosome 4H
B.P. Forster
Division of
Genetics, Scottish Crop Research Institute
Invergowrie,
Dundee DD2 5DA, UK
The
past ten years have seen genetic mapping in barley shift from largely
morphological marker maps to molecular marker maps. Molecular marker maps,
especially those based on PCR, continue to be published. Mapped markers however
are being used increasingly to locate genes of interest including quantitative traits.
Different classes of molecular markers are often combined in targeting and
locating genes of interest. Future mapping exercises are expected to focus on
functional genes and their associated flanking markers.
The
development of molecular markers has contributed to increased activity in
genetic studies of barley in the last year. Published papers involve new
marker, gene and QTL maps based on recombination, physical mapping and
comparative mapping. A brief summary of work I am aware of on chromosome 4H is
given below.
MARKER MAPPING
Marker
maps of barley include RFLPs, RAPDs, AFLPs, SSRs, S-SAPs and BARE-1.
Established marker maps include those developed in Australia, England and North
America (based on RFLPs), The Netherlands (based on AFLPs), Denmark and Germany
(RAPDs) and in the USA and Scotland (SSRs). Regularly up-dated web-sites have
been set up for some of these. New marker maps include PCR derivatives of
established markers. In Japan, a sequence-tagged site (STS) map of barley has
been further developed by Mano et al.,
1999, the published map shows 10 linked STSs on 4H covering about 57 cM. The
PCR-based STSs were developed from terminal sequences of cloned RFLPs. A barley
genetic map composed of random amplified microsatellite polymorphisms (RAMPs)
has been developed in Spain by Dávila et
al., 1999. Telomere-associated sequences have been used to map 10 out of 14
telomeres, including both ends of 4H (Kilian et al., 1999). The map distance between the two 4H telomeres, Tel4S and Tel4L, was relatively short at 150 cM. On this evidence 4H may be
the shortest barley chromosome in terms of its recombination map
TRAIT MAPPING
The development of high-density molecular marker maps
has been used to identify, locate and quantify QTLs determining a number of
traits including economically important traits such as yield, quality and
disease resistance.
Biotic stress
A
primary QTL for net blotch resistance was detected in a spring barley cross in
the region of mlo on the long arm of
4H (Thomas, 1999). Two QTLs for partial resistance to leaf rust mapped to 4H
(Qi et al., 1999). Resistance genes
have been mapped in rice and barley by Leister et al., 1999, with Hv-b32
mapping to 4H (Ym-11, resistance to
yellow mosaic virus, is also indicated on 4H in Leister et al., 1999).
Abiotic stress
Four
chromosomal regions have been identified for boron tolerance. One on 4H was
associated with boron uptake, root length response, dry matter production and
symptom expression (Jefferies et al.,
1999). Dehydrin genes, which are related to low temperature and dehydration
responses, have been located using wheat/barley chromosome addition lines. One
of eleven dehydrin genes, Dhn6, was
located to 4H (Choi et al., 1999).
Other
work on wheat/barley chromosome addition lines indicated that 4HL carries a
factor concerned with seedling salt tolerance (expressed as dry weight and ð13C,
Ellis et al., 2000).
Physiological
traits
A
gibberellic acid-insensitive dwarfing gene (Dwf2)
has been located on 4H using RFLPs and SSRs in an F2 population
(Ivandic et al., 1999). QTLs analysis
in conjunction with AFLPs has been used to locate QTLs determining yield. Of
the nine traits studied two, i.e. specific leaf area at flowering and yield had
QTLs on 4H (Yin et al., 1999).
Favourable QTL alleles for grain yield have been identified on all seven barley
chromosomes in the spring barley cross, Steptoe x Morex (Zhu et al., 1999).
Sequence analysis
Michalek
et al., 1999 compared partial
sequences of mapped cDNA and genomic clones with data base sequences of
proteins and nucleic acids. Putative identifications were made based on
sequence similarity. For chromosome 4H these included: two sulphate
transporters, a light-inducible protein (ELIP) and NADH dehydrogenase. The
sequence data can be exploited in converting RFLPs into STS (PCR-based)
markers.
Others
Larson
and Raboy, 1999 using wheat/barley chromosome addition lines mapped a myo-inositol 1-phosphate synthase (MIPS)
gene to 4H. A hairy leaf sheath gene, Hsb
derived from Hordeum bulbosum
was mapped on 4HL using RFLPs (Korzun et
al., 1999). A root fluorescence mutant, frp
( fluorescent pink reaction) produced by gamma-ray treated was located close to
glf3 (glossy leaf 3) on 4H (Takeda
and Chang, 1998).
COMPARATIVE
MAPPING
Some
genetic markers, notably RFLPs but not SSRs, can be used in comparative studies
of homologous and homoeologous chromosomes. Comparisons of 4H maps include:
gene maps, RFLP maps with QTLs (agronomic, stress, developmental traits), AFLP
maps and a wheat group 4 map (Forster et
al., 2000). Barley/wheat comparisons have been made among 4HS v 4BS v 4DS
with reference to Dwf2 v Rht-B1 v Rht-D1 and RFLP markers (Ivandic et al., 1999). Work on MIPS has involved comparisons of maize 1S
and group 4 homoeologues of the Triticeae
(Larson and Raboy, 1999). Recombination break points between 4H of H. vulgare and H. bulbosum and comparisons with 5R of rye have been made using
RFLPs linked to hairy leaf sheath (Korzun et
al., 1999).
PHYSICAL
MAPPING
The
physical locations of rDNA sequences were examined by in situ hybridisation by Taketa et
al., 1999. A minor site for 18-25S rDNA was located on the short arm of 4H.
ORDERLY
ARRANGEMENT OF BARLEY CHROMOSOMES
Linde-Laursen
and Bothmer, 1999 studied the order of elimination of barley chromosomes in
hybrids with Hordeum lechleri. In one
cross 4H was the last to be eliminated, but varietal differences were found.
References
Choi D.-W., B. Zhu and T.J. Close. 1999. The barley (Hordeum vulgare L.) dehydrin multigene
family: sequences, allele types, chromosome assignments, and expression
characteristics of 11 Dhn genes of cv
Dicktoo. Theoretical and Applied Genetics 98: 1234-1247.
Dávila J.A., Y.Loarce and E. Ferrer. 1999. Molecular characterization
and genetic mapping of random amplified microsatellite polymorphism in barley.
Theoretical and Applied Genetics 98: 265-273.
Ellis R.P., B.P. Forster, D. Robinson, L.L. Handley, D.C. Gordon, J.R.
Russell and W. Powell. 2000. Wild barley: a source of genes for crop
improvement in the 21st century? Journal of Experimental Botany 51 (in press).
Forster B.P., R.P. Ellis, W.T.B. Thomas, A.C. Newton, R. Tuberosa, D.
This, R.A. El-Enein, M.H. Bahri and M. Ben Salem. 2000. The development and
application of molecular markers for abiotic stress tolerance in barley.
Journal of Experimental Botany 51 (in press).
Ivandic V., R.A. Malyshev, V. Korzum, A. Graner and A. Borner. 1999.
Comparative mapping of gibberellic acid-insensitive dwarfing gene (Dwf2) on
chromosome 4HS in barley. Theoretical and Applied Genetics 98: 728-731.
Jefferies S.P.,
A.R. Barr, A. Karakousis, J.M. Kretschmer, S. Manning, K.J. Chalmers, J.C.
Nelson, A.K.M.R. Islam and P. Langridge P. 1999. Mapping of chromosome regions
conferring boron toxicity tolerance in barley (Hordeum vulgare L.). Theoretical and Applied Genetics 98:
1293-1303.
Kilian A. and A.
Kleinhofs. 1992. Cloning and mapping of telomere-associated sequences from
Hordeum vulgare L. Molecular and General Genetics 235: 153-156.
Kilian A., D.
Kudrna and A. Kleinhofs. 1999. Genetic and molecular characterization of barley
chromosome telomeres. Genome 42: 412-419.
Korzun V., R.A.
Malyshev, R.A. Pickering and A. Börner. 1999. RFLP mapping of a gene for hairy
leaf sheath using a recombinant line from Hordeum
vulgare L. x Hordeum bulbosum L. cross. Genome 42: 960-963.
Larson S.R. and
V. Raboy. 1999. Linkage mapping of maize and barley myo-inositol 1-phosphate synthase DNA sequences: correspondence
with a low phytic acid mutation. Theoretical and Applied Genetics 99: 27-36.
Leister D., J. Kurth,
D.A. Laurie, M. Yano, T. Sasaki, A. Graner and P. Schulze-Lefert. 1999. RFLP-
and physical mapping of resistance gene homologues in rice (O. sativa) and barley (H. vulgare). Theoretical and Applied
Genetics 98: 509-520.
Linde-Laursen I.
and R. von Bothmer. 1999. Orderly arrangement of the chromosomes within barley
genomes of chromosome-eliminating Hordeum
lechleri x barley hybrids. Genome 42: 225-236.
Mano Y., B.E.
Sayed-Tabatabaei, A. Graner, T. Blake, F. Takaiwa, S. Oka and T. Komatsuda.
1999. Map construction of sequence-tagged sites (STSs) in barley (Hordeum vulgare L.). Theoretical and
Applied Genetics 98: 937-946.
Michalek W., G.
Künzel and A. Graner. 1999. Sequence
analysis and gene identification in a set of mapped RFLP markers in barley (Hordeum vulgare). Genome 42: 849-853.
Qi X., G. Jiang,
W. Chen, R.E. Niks, P. Stam and P. Lindhout. 1999. Isolate-specific QTLs for
partial resistance to Puccinia hordei
in barley. Theoretical and Applied Genetics 99: 877-884.
Takeda K. and
C.L. Chang. 1998. Studies on root fluorescent mutants in barley. Bulletin of
the Research Institute for Bioresources, Okayama, Japan 5: 193-202.
Taketa S., G.E.
Harrison and J.S. Heslop-Harrison. 1999. Comparative physical mapping of the 5S
and 18-25S rDNA in nine wild Hordeum
species and cytotypes. Theoretical and Applied Genetics 98: 1-9.
Thomas W.T.B.
1999. QTL mapping of Net Blotch resistance in a spring barley cross. Proceedings of a Workshop on “Disease resistance and cereal leaf
pathogens beyond the year 2000”. COST Action 817 “Population studies of
airborne pathogens in cereals”. Martina Franca, Italy, p.54-55.
Yin Z., P. Stam,
C. Johan Dourleijn and M.J. Kropff. 1999. AFLP mapping of quantitative trait
loci for yield-determining physiological characters in spring barley.
Theoretical and Applied Genetics 99: 244-253.
Zhu H., G.
Briceño, R. Dovel, P.M. Hayes, B.H. Liu, C.T. Liu and S.E. Ullrich. 1999.
Molecular breeding for grain yield in barley: an evaluation of QTL effects in a
spring barley cross. Theoretical and Applied Genetics 98: 772-779.
Coordinators report: Chromosome 5H (7)
George
Fedak
Eastern
Cereal and Oilseed Research Centre
Agriculture
& Agri-Food Canada
Ottawa,
Ontario
Canada
K1A 0C6
De la Pena et al., 1999 were the first group to
publish on the assignment of molecular markers to QTL for components of
Fusarium head blight resistance in barley. Ninety-four previously mapped RFLP
markers were screened across a mapping population of 101 F4:7 lines obtained
from crossing Chevron and M69. A total of 10 QTL were detected for FHB
symptoms, 11 for DON accumulation and 4 for kernel discoloration. Chromosome 7
(5H) contained one QTL for each of the above traits plus a plant height QTL. A
heading data QTL on chromosome 7 coincided with one for kernel discoloration.
A high density AFLP map obtained from 103RILs from a 94 x Vada (resistant) hybrid was used to identify QTL for partial resistance to leaf rust (Qi et al., 1998; 1999). The population was phenotyped at both the seedling and adult plant stages with isolate 1.2.1. A total of 6 QTLs for partial resistance to leaf rust were identified in the population. Three of the six QTL were effective at the seedling stage and six were effective at the adult plant stage indicating that two of them were effective at both stages. The QTL designated as Rpq4 was located at a distal location on the short arm of chromosome 7 (5H), was effective at the adult plant stage and combined with Rphq3 on chromosome 6 accounted for most of the phenotypic variance at the adult plant stage. They point out that the map position on chromosome 7S (5HS) of Rpq4 does not coincide with, nor is it close to the location of previously mapped race-specific genes on that chromosome. For example Rph9 and Rph12 (which have been shown to be allelic) have been mapped to the distal portion of the long arm of chromosome 7 (Borovkova et al., 1997). An additional
race specific gene, RphQ (a presumed allele at the Rph2
locus) has been mapped near the centromere of 7S and quite distant from Rpq4 at the distal end of that
chromosome.
The same L94 x
Vada RIL population was subsequently inoculated with a different isolate,
isolate 24 (Qi et al., 1999). An
additional QTL for seedling resistance (Rphq7)
was detected and mapped to a distal location on the long arm of chromosome 7.
In summary, of
the total of 8 QTL for partial resistance to leaf rust at the adult stage
identified in the two studies, three were effective against both isolates and
five were effective against only one of the two isolates. Only one QTL had a
substantive effect at both seedling and adult plant stages.
Dehydrins
are LEA proteins that are induced in response to low temperature, drought and
salinity. Cumulative studies (Choi et al.,
1999) indicate that the barley genome contains 13 Dhn genes, based on screening of existing genomic libraries. They
are located on chromosomes 3H, 4H, 5H and 6H. Chromosome 5H contains the loci Dhn1, Dhn2 and Dhn9.
The
three dehydrin (Dhn) genes on
chromosome 5H span a 20 cM region. Included in this interval is Sgh2 the gene determining the
winter/spring growth habit (Choi et al.,
1999).
A new
cereal cyst nematode (Heterodosa avenae)
resistance gene, designated as Rha4,
from the Australian barley cultivar Galleon was mapped to chromosome 5H (Barr et al., 1998). It is flanked by the RFLP
markers XYL (6.2 cM) and BCD298 (12.5 cM).
Three
QTL for partial resistance to bacterial leaf streak (Xanthomonas campestris pv. hordei)
were found in the Steptoe x Morex DH population (El Attari et al., 1998). Two were mapped on chromosome 3H and one on 5H near
the marker ABC155. The three QTL accounted for 30% of the phenotypic variation
in the population.
Severity of five
diseases under conditions of natural infection at various test sites was
evaluated in the Harrington/TR306 DH population (Spaner et al., 1998). QTL for resistance to three of these diseases
involved chromosome 5H. QTL determining resistance to powdery mildew were
located on chromosome 4H and 5H; for resistance to leaf rust on chromosomes 2H,
5H and 6H and QTL for resistance to net blotch were identified on chromosomes
4H, 5H, 6H and 7H.
Druka et al., 1999 have done some fine-scale
mapping around the rpg4 locus located
in the subtelomeric region of chromosome 5HL. The distal RFLP markers around
this locus are ABG391 and R273 at 1.6 and 0.8 cM, respectively. The proximal
RFLP marker is Aga5 at about 1.8 cM. In preparation for map-based cloning, new
RFLP markers have been found that bracket that locus and spaced about 0.5 cM
apart. A barley BAC contig across the rpg4
region has been established.
An
adult plant resistance QTL for stripe rust resistance in the cultivars
Cali-sile and CMB643 has been mapped onto chromosome 5H (Hayes et al., 1999).
Seah et al., 1998 amplified a number of resistance gene analogs (RGA) from wheat and barley using specific primers derived from conserved sequences of the Cre3 cereal cyst nematode resistance locus. Amplifications were made from barley cultivars Chebec and Harrington and mapping of RGA was done on the Steptoe x Morex and Chebec x Harrington DH populations. Two of the most divergent barley RGAs (from the 17 isolated) were mapped. By virtue of the multiple banding patterns it was concluded that the RGAs belonged to multigene families. The clones mapped mainly to long arms of chromosomes 2H(2), 5H(7) and 7H(1). The RGAs mapped to at least seven non homologous loci and in many cases were linked to known disease
resistance loci. On chromosome 5H two and possibly
three independent RGA loci were identified.
References
Barr, A.R., K.J.
Chalmers, A. Karakousis, J.M. Kretschmer, S. Manning, R.C.M. Lance, J. Lewis,
S.P. Jeffries and P. Langridge. 1998. RFLP mapping of a new cereal cyst
nematode resistance locus in barley. Plant Breed. 117: 185-187.
Borovkova, I.G., Y. Jin,
B.J. Steffenson, A. Kilian, T.K. Blake and A. Kleinhofs. 1997. Identification
and mapping of a leaf rust resistance gene in barley line Q21861. Genome
40:236-241.
Choi, D.W., B. Zhu and
T.J. Close. 1999. The barley (Hordeum
vulgare L.) dehydrin gene family; sequences, allele types chromosome
assignments, and expression characteristics of 11 Dhn genes of cv. Dicktoo. Theor. Appl. Genet. 98: 1234-1247.
Choi, D.-W., Koag, M.C.
and T.J. Close. 1999. Allelic variation and genetic mapping of barley dehydrin
(Dhn) genes. Plant and Animal Genome VII abstract p. 182.
De la Pena, R.C., K.P.
Smith, F. Capettini, G.L. Muehlbauer, M. Gallo-Meagher, R. Dill-Macky, D.A.
Somers and D.C. Rasmussen. 1999. Quantitative trait loci associated with
resistance to Fusarium head blight and kernel discoloration in barley. Theor.
Appl. Genet. 99: 561-569.
Druka, A., D. Kudrna, F.
Han, A. Kilian, B. Steffenson, Y. Yo, D. Frisch, J. Tomkins, R. Wing and A.
Kleinhofs. 1999. Map based cloning of barley rpg4 gene. Plant and Animal Genome VII, abstract p. 178.
El Attari, H., A. Rebai,
P.M. Hayes, G. Barrault, G. Dechamp-Guillaume and A. Sarrafi. 1998. Potential
of double haploid lines and localization of quantitative trait loci (QTL) for
partial resistance to bacterial leaf streak (Xanthomonas campestris pv hordei)
in barley. Theor. Appl. Genet. 96: 95-100.
Hayes, P.M., X.
Chen, A. Corey, M. Johnston, A. Kleinhofs, J. Korte, D. Kudrna, T. Toojinda and
H. Vivar. 1999. A summary of barley stripe rust mapping efforts. Plant and
Animal Genome VII p. 179.
Qi, X., R.E. Niks, P.
Stam and P. Lindhout. 1998. Identification of QTL for partial resistance to
leaf rust (Puccinia hordei) in
barley. Theor. Appl. Genet. 96: 1205-1215.
Qi, X., G. Jiang, W.
Chen, R.E. Niks, P. Stam and P. Lindhout. 1999. Isolate specific QTL for
partial resistance to Puccinia hordei
in barley. Theor. Appl. Genet. 99: 877-884.
Seah, S., K.
Sivasithamparam, A. Karakousis and E.S. Lagudah. 1998. Cloning and
characterization of a family of disease resistance gene analogs from wheat and
barley. Theor. Appl. Genet. 97: 937-945.
Spaner, D., L.P. Shugar,
T.M. Choo, I. Falak, K.G. Briggs, W.G. Legge, D.E. Falk, S.E. Ullrich, N.A.
Tinker, B.J. Steffenson and D.E. Mather. 1998. Mapping of disease resistance
loci in barley on the basis of visual assessment of naturally occurring
symptoms. Crop Sci. 38: 843-850.
Coordinator's
Report: Chromosome 7H
Lynn
Dahleen
USDA-Agricultural
Research Service,
Fargo,
ND 58105, USA
Mapping projects continued at full speed
in 1999, adding many new markers, genes and QTLs to all chromosomes of barley,
including chromosome 7H (1). This report briefly describes the additions to
chromosome 7H (1), covering all the literature I was able to find and obtain
through North Dakota State University’s library. If you have additional papers
with markers or genes on chromosome 7H (1), please mail a reprint to me for
inclusion in next years report.
Kilian et al.,.1999 continued their work on barley telomere markers,
increasing the number of marked chromosome ends to ten. They identified
additional markers that cosegregate with the most distal marker on 7HS (1S), Tel1S. These new markers are PCR-based
using a single primer encoding HvRT-like sequences. These markers should be
usable in any lab as they were reproducible when different temperature profiles
and Taq polymerases were used.
Unfortunately, they were unable to locate a telomeric marker for four of the
chromosome arms, including the long arm of chromosome 7H (1).
New types of PCR markers were developed
by Dávila et al., 1999, based on
microsatellite primed-PCR (MP-PCR), which uses a dinucleotide repeat primer
with an arbitrary 10-mer primer, and random amplified microsatellite
polymorphisms (RAMP), which uses a single dinucleotide repeat primer to amplify
DNA. Both of these methods amplify multiple bands. Thirty-three of these bands
were mapped on the Steptoe/Morex map, six of them on chromosome 7H (1). These
included two markers that mapped to the end of the short arm, one in the middle
of the short arm, two on the short arm near the centromere, and one on the long
arm near the centromere. Several of these markers mapped to regions with poor
RFLP coverage.
Leister et al., 1999 examined consensus resistance gene nucleotide binding
site-leucine-rich repeat (NBS-LRR) homolog sequence diversity and conservation,
and compared linkage of these sequences with mapped resistance genes in rice
and barley. Out of the nine barley NBS-LRR homologs evaluated, one mapped to
the short arm of chromosome 7H (1). The mapping populations tested did not
segregate for Rpg1, but comparative
placement based on flanking markers indicated that the NBS-LRR homolog mapped
to the same region as Rpg1.
Shan et
al., 1999 identified 174 chromosome-specific AFLP markers in wheat-barley
addition lines, including 40 located on barley chromosome 7H (1). They then
tried to convert 10 barley-specific and 16 wheat-specific AFLP fragments to
sequence-specific PCR markers. Only six of the 26 primer sets amplified DNA
only from the expected chromosome, indicating that there often are problems in
converting AFLP polymorphisms into sequence-specific PCR markers.
Mano et
al., 1999 developed 43 new STS primer pairs from sequences of RFLP clones,
including 19 that amplified polymorphic products between Azumamugi (A) and
Kanto Nakate Gold (KNG). Four of the STS products located on chromosome 7H (1)
by wheat-barley addition lines were monomorphic on A x KNG, preventing precise
map location. Of the other three primer sets, MWG832 showed polymorphism after
amplification, and cMWG704 and MWG2062 detected polymorphism after restriction
enzyme digestion. The chromosome locations of these STS markers matched the
locations of the RFLP clones that were used to determine the primer sequences.
Ramsay et al., 1999 examined 290 clones [four from chromosome 7H (1)],
containing dinucleotide repeats from SSR-enriched libraries. A high percentage
of the sequences were associated with retrotransposon-like and other repetitive
elements. One of the chromosome 7H (1) clones, Bmag0004, was directly
associated with the retrotransposon BARE-1. The other three clones, Bmag0110,
Bmag0217 and Bmag0228 showed significant homology to the dispersed repetitive
element R173. The specific relationship of the SSR sequences to the
retrotransposon sequences was determined and characteristic patterns of SSR
sequence insertion were observed for each retrotransposon. The authors discuss
some of the implications of these associations.
Partial sequences of 43 cDNA and 259
genomic DNA clones were determined by Michalek et al., 1999 and compared to sequences on the major protein and
nucleic acid databases. For the cDNA clones, 53% showed significant similarity
to protein sequences and 35% to nucleic acid sequences. Only 9% of the genomic
clones showed similarities to nucleic acid sequences. From chromosome 7H (1),
six of the cDNA clones and three of the genomic clones sequenced showed
similarity to known genes in the databases. Types of genes showing similarity
to the chromosome 7H (1) clones included an AT-hook protein, a transketolase,
and a peroxidase. The nine genes showing similarity came from a variety of
different plant and bacterial species.
Ma et
al., 1999 have been examining genes involved in phytosiderophore
biosynthesis. These mugineic acids are involved in Fe acquisition, helping
barley survive in soils with low available iron. Two genes involved in
hydroxylations that convert 2'-deoxymugineic acid to 3-epihydroxymugineic acid
(epiHMA) work in concert, one on chromosome 4H that hydroxylates the C-2'
position, and one on the long arm of chromosome 7H (1), which hydroxylates the
C-3 position. Both genes are required to produce epiHMA.
A major gene conferring resistance to the
spot form of net blotch (SFNB) was identified and mapped from the cultivar
Galleon by Williams et al., 1999.
This gene, named Rpt4, explained 80%
of the variation in the Galleon x Haruna Nijo mapping population, and was
located on the long arm of chromosome 7H (1). When the nearest linked RFLP
marker (PSR117-D) was used to select resistant lines from another
Galleon-derived population, the SFNB phenotype was correctly predicted in 22
out of 24 progeny, verifying the effectiveness of the linked marker for
marker-assisted selection of Rpt4.
Qi et
al., 1999 located isolate-specific QTLs involved in partial resistance to
barley leaf rust at both the seedling and adult plant stages. Three of these
QTL were located on chromosome 7H (1). Rphq1,
which had been found previously to have a small effect against isolate 1.2.1,
had no effect on isolate 24. The other two QTLs from chromosome 7H (1), Rphq8 and Rphq9, were only effective against isolate 24, only at the adult
plant stage. The map position for Rphq8
coincided with a minor QTL for days to heading.
Stripe rust reaction was evaluated by
Pecchioni et al., 1999 in 97 doubled
haploid lines from Proctor x Nudinka, and they placed seven pathogen response
(PR) genes on the linkage map. Two of the PR genes cosegregated near the end of
the long arm of chromosome 7H (1), Chi1 coding
for barley chitinase and Tha, coding
for a barley thaumatin-like protein. A major QTL (r2=58.5%) was
located near the centromere on chromosome 7H (1). A second, minor QTL (r2=3.0%)
was located in the middle of the long arm of chromosome 7H (1). None of the
QTLs identified were located near the PR genes tested.
Therrien, 1999 reevaluated a data set
from a population segregating for loose smut resistance and a number of
morphological markers. What was first thought to be a two-gene system for
resistance, was in fact a quantitative system. Four of the morphological
markers showed significant linkage including Chlorina seedling 8 (fch8) on chromosome 7H (1), which was
linked (44 cM) to resistance.
Yin et
al., 1999 used an AFLP map to locate QTL for yield-determining physiological
traits in a Prisma x Apex recombinant inbred population. A threshold LOD value
of 3.0 was used to select QTL. Regions with an LOD between 2.5 and 3.0 were
considered “suggestive” for QTLs. Of the eight QTL located on chromosome 7H
(1), six were suggestive for QTLs for height, pre- and post-flowering duration
and fraction of shoot biomass partitioned to ears. Two QTL from chromosome 7H
(1) had an LOD>3.0, one associated with the fraction of shoot biomass
partitioned to leaves, and the other associated with yield. All eight of these
QTL were detected in only one of the two years of field tests.
Kleinhofs, 1999 provided an update on the
current state of the integrated molecular/morphological map.
Morphological/physiological markers placed on the chromosome 7H (1) molecular
map include brh1, wax, fch12,
cer-ze, fch5, Lga, nar3, pmr, nud, mo6b, lks2, msg10, and Xnt1.
In another map integration paper, Korzun
and Künzel, 1999 report on the completion of their project to integrate the
physical and molecular maps of barley by integrating 32 translocation
breakpoints of chromosome 7H (1) into the corresponding Igri/Franka-derived
RFLP map. Seven of the translocation breakpoints mapped between cosegregating
markers, providing a physical split in marker clusters. This project provides
barley researchers with valuable materials for gene mapping and isolation.
References
Dávila,
J. A., Y. Loarce and E. Ferrer. 1999. Molecular characterization and genetic
mapping of random amplified microsatellite polymorphism in barley. Theor. Appl.
Genet. 98:265-273.
Kilian,
A., D. Kudrna and A. Kleinhofs. 1999. Genetic and molecular characterization of
barley chromosome telomeres. Genome 42:412-419.
Kleinhofs,
A. 1999. Coordinator’s report: integrating barley molecular and morphological/
physiological marker maps. BGN 29:
Korzun,
L. and G. Künzel. 1999. Integration of 32 translocation breakpoints of
chromosome 7H (1) into the corresponding Igri/Franka-derived RFLP map. BGN 29:
Leister,
D., J. Kurth, D.A. Laurie, M. Yano, T. Sasaki, A. Graner and P.
Schulze-Lefert.1999. RFLP- and physical mapping of resistance gene homologues
in rice (O. sativa) and barley (H. vulgare). Theor. Appl. Genet.
98:509-520.
Ma,
J.F., S. Taketa, Y.-C. Chang, T. Iwashita, H. Matsumoto, K. Takeda and K.
Nomoto. 1999. Genes controlling hydroxylations of phytosiderophores are located
on different chromosomes of barley (Hordeum
vulgare L.). Planta 207:590-596.
Mano,
Y., B.E. Sayed-Tabatabaei, A. Graner, T. Blake, F. Takaiwa, S. Oka and T.
Komatsuda. 1999. Map construction of sequence-tagged sites (STSs) in barley (Hordeum vulgare L.). Theor. Appl. Genet.
98:937-946.
Michalek,
W., G. Künzel and A. Graner. 1999. Sequence analysis and gene identification in
a set of mapped RFLP markers in barley (Hordeum
vulgare). Genome 42:849-853.
Pecchioni,
N., G. Vale, H. Toubia-Rahme, P. Faccioli, V. Terzi and G. Delogu. 1999.
Barley-Pyrenophora graminea interaction: QTL analysis and
gene mapping. Plant Breeding 118:29-35.
Qi,
X., G. Jiang, W. Chen, R.E. Nicks, P. Stam and P. Lindhout. 1999.
Isolate-specific QTLs for partial resistance to Puccinia hordei in barley. Theor. Appl. Genet. 99:877-884.
Ramsay,
L., M. Macaulay, L. Cardle, M. Morgante, S. degli Ivanissevich, E. Maestri, W.
Powell and R. Waugh. 1999. Intimate association of microsatellite repeats with
retrotransposons and other dispersed repetitive elements in barley. Plant J.
17:415-425.
Shan,
X., T.K. Blake and L.E. Talbert. 1999. Conversion of AFLP markers to
sequence-specific PCR markers in barley and wheat. Theor. Appl. Genet.
98:1072-1078.
Therrien,
M.C. 1999. The possibility of quantitative inheritance of loose smut in barley.
BGN 29:
Williams,
K.J., A. Lichon, P. Gianquitto, J.M. Kretschmer, A. Karakousis, S. Manning, P.
Langridge and H. Wallwork. 1999. Identification and mapping of a gene
conferring resistance to the spot form of net blotch (Pyrenophora teres f maculata)
in barley. Theor. Appl. Genet. 99:323-327.
Yin,
X., P. Stam, C. Johan Dourleijn and M.J. Kropff. 1999. AFLP mapping of
quantitative trait loci for yield-determining physiological characters in
spring barley. Theor. Appl. Genet. 99:244-253.
Integrating Molecular and
Morphological/Physiological Marker Maps
A. Kleinhofs
Dept. Crop and Soil Sciences
Washington State University
Pullman, WA 99164-6420, USA
Considerable
progress in refining barley genetic maps has taken place during the past year.
I have posted the BIN maps at “http://barleygenomics.wsu.edu”.
These maps are based on the Steptoe x Morex (SM) map and markers mapped in
other crosses are located only to BINs unless they can be approximated
precisely to the SM map based on nearby common markers. These maps are
frequently updated as new information is incorporated. However, they are not
very user friendly and it is often hard to find the information one is looking
for, therefore, we are in the process of developing a searchable database that
is connected to the maps. A test version is available at “http://www.wsu.edu/~druka/webdata/search.htm”.
Comments and suggestions for improvement are welcome. Barley morphological
modeling can be found at
“http://mansfeld.ipk-gatersleben.de/bucksorlin/”
and
Künzel’s physical maps at “http://wheat.pw.usda.gov/ggpages/Barley_physical/”.
The
Vrs1 locus was mapped to chromosome 2
(2H) with 929 chromosomes 0.1 cM distal from cMWG669 and 0.9cM proximal from
MWG865 (Komatsuda et al., 1999). The MWG865 marker is mapped in SM allowing accurate
positioning of the Vrs1 locus in the
chr2 BIN map. Since the Vrs1 locus
has been used to map other morphological markers (Franckowiak, 1997; Sogaard and von
Wettstein-Knowles, 1987) it allows reasonable positioning of these markers as well.
Three
dwarfing genes were mapped with reference to molecular markers (Borner et al., 1999; Ivandic et al.,
1999). The Dwarf2 (Dwf2) gene was mapped to chromosome 4S
(4HS) 5.7 cM proximal to hvOle (PCR
derived from the Ole1 locus which is
mapped in SM). The GA-sensitive (GA-less;
gal) mutant was mapped to chromosome
2 (2H) 3.6 cM distal from the cosegregating markers MWG581, MWG882, and MWG2212
and 9.5 cM proximal to ksuG5. On the SM map the closest flanking markers are
MWG503, 6.7 cM proximal and ksuD22, 12.5 cM distal. The GA-insensitive (GA-ins; gai) mutant cosegregated with MWG2058 and MWG2287 on chromosome 2
(2H) (not on the SM map) and was 1.8 cM proximal to MWG557 (cosegregates with
B15C in the SM map). These markers allow the placement of all 3 dwarfing loci
on the combined BIN maps. A high resolution map of the brh1 (brachytic1) mutant
FN53 induced in cv. Steptoe placed it less than 1 cM distal to BCD129 on
chromosome 1 (7H) (Li Ming et al, AK
lab, unpublished). These data are in good agreement with those previously
reported (Jin et
al., 1993) and allow us to position fch12 (fc) and fch5 (f5).
Two
low phytic acid (lpa1-1 and lpa2-1) mutants were mapped to barley
chromosomes 2 (2H) and 1 (7H), respectively (Larson et al., 1998). The lpa1-1 locus
cosegregated with ABC157 and was placed on the map. The lpa2-1 locus resides in a poorly defined region distal from ABC151B
and proximal to ABC310B and was not placed on the map. The barley myo-inositol
1-phosphate synthase gene was cloned and mapped to a single locus on chromosome
4 (4H), thus, neither lpa1 nor lpa2 seem to represent the structural
gene for this enzyme (Larson and Raboy, 1999).
The
Brittle rachis 2 (Btr2) trait was mapped to chromosome 3
(3H) between the markers ABC171 and ABG396 using the Bowman backcrossed line
GSHO1937 obtained from J. Franckowiak (Kandemir et al., 2000). The ABC171-ABG396 segment is approximately 38 cM and
approximates the H. spontaneum region
introgressed into cv. Bowman. This locus was not placed on the map.
Magnesium
chelatase subunits are encoded by the Xantha-f
(xan-f), Xantha-g (xan-g), and Xantha-h
(xan-h) genes (Jensen et al., 1996). The genes were mapped to chromosome arm location using
barley-wheat telosomic addition lines. xan-f
mapped to 2S while xan-h mapped to 1
(7H)L (Hansson et al., 1998). xan-g was
recently cloned, but has not been mapped (Petersen et al., 1999). The gene designations used do not correspond to those in
BGN vol. 26.
The
updated morphological marker maps are shown in Fig.1 (following page).
References
Borner,
A., V. Korzun, S. Malyshev, V. Ivandic, V. and A.
Graner. 1999. Molecular mapping of two dwarfing genes differing in their GA
response on chromosome 2H in barley. Theor. Appl. Genet. 99: 670-675.
Franckowiak,
J. 1997. Revised linkage maps for morphological markers in barley, Hordeum vulgare. Barley Genetics
Newsletter, 26: 9-21.
Hansson,
M., S.P. Gough, C.G. Kannangara and D. von Wettstein. 1998. Chromosomal
locations of six barley genes encoding enzymes of chlorophyll and heme
biosynthesis and the sequence of the ferrochelatase gene identify two
regulatory genes. Plant Physiol. Biochem. 36: 545-554.
Ivandic,
V., S. Malyshev, V. Korzun, A. Graner and A. Borner. 1999. Comparative mapping
of a gibberellic acid-insensitive dwarfing gene (Dwf2) on chromosome 4HS in
barley. Theor. Appl. Genet. 98: 728-731.
Jensen,
P. E., R.D. Willows, B.L. Petersen, U.C. Vothknetcht, B.M. Stumman, C.G.
Kannangara, D. von Wettstein and K.W. Henningsen. 1996. Structural genes for
Mg-chelatase subunits in barley: Xantha-f, -g and -h. Mol. Gen. Genet. 250: 383-394.
Jin, Y.,
B.J. Steffenson and J.D. Franckowiak. 1993. Linkage between the Rpg1 gene for
stem rust resistance and the f5 locus on barley chromosome 1. Crop Sci. 33:
642-643.
Kandemir,
N., D.A. Kudrna, S.E. Ullrich and A. Kleinhofs. 2000. Molecular marker assisted
genetic analysis of head shattering in six-rowed barley. Theor. Appl. Genet.
(in press).
Komatsuda,
T., W. Li, F. Takaiwa and S. Oka. (1999). High resolution map around the vrs1 locus controlling two- and
six-rowed spike in barley, Hordeum
vulgare. Genome 42: 248-253.
Larson,
S. R. and V. Raboy. 1999. Linkage mapping of maize and barley myo-inositol
1-phosphate synthase DNA sequences: correspondence with low phytic acid
mutation. Theor. Appl. Genet. 99: 27-36.
Larson,
S. R., K.A. Young, A. Cook, T.K. Blake and V. Raboy. 1998. Linkage mapping of
two mutations that reduce phytic acid content of barley grain. Theor. Appl.
Genet. 97: 141-146.
Petersen,
B. L., M.G. Moller, P.E. Jensen and K.W. Henningsen. 1999. Identification of
the Xan-g gene and expression of the
Mg-chelatase encoding genes Xan-f, -g,
and -h in mutant and wild type barley
(Hordeum vulgare L.). Hereditas 131:
165-170.
Søgaard,
B. and P. von Wettstein-Knowles. 1987. Barley: genes and chromosomes. Carlsberg
Res. Commun. 52: 123-196.
Coordinator's Report:
Barley Genetics Stock Collection
A.Hang
USDA-ARS, National Small Grains Germplasm
Research Facility,
Aberdeen, Idaho 83210, USA
More
than 400 barley genetic stocks derived from Bowman backcrosses were planted in
the field at Aberdeen, Idaho, for evaluation and for seed increase.
A
set of 94 doubled haploid (DH) multiple markers genetic stocks, designated
Oregon Wolfe Barley (OWB) and developed by Dr. Patrick Hayes at Oregon State
University, were planted in Aberdeen, Idaho for seed increase. Seeds are
maintained at the National Small Grains Germplasm Research Facility at Aberdeen
for distribution.
Seeds
of the following multiple marker stocks for the seven barley chromosomes were
also increased:
Markers
for chromosome 1H: Third outer glume, Chlorina seedling 3 (trd, fch3)
Markers
for chromosome 2H: White streak 4, Glossy sheath 5 (wst4, gsh5)
Markers for
chromosome 3H: Curly 2, Chlorina seedling 2 (cur2, fch2); Curly 2, Yellow streak 2, Uzu or Semi-brachytic (cur2, yst2, uzu)
Markers
for chromosome 4H: Chlorina seedling 9, Hooded lemma, Short awn 5 (fch9, Kap, lks5)
Markers
for chromosome 5H: Variegated 1, Mottled leaf 2 (var1, mtt2)
Markers
for chromosome 6H: Uniculm 2, Orange lemma (cul2,
rob)
Markers for
chromosome 7H: Chlorina seedling 8, Short awn 2, Naked caryopsis (fch8, lks2, nud) Glossy sheath 3,
Chlorina seedling 12 (gsh3, fch12).
The
world collection of translocation stocks in barley collected by Dr. R.T. Ramage
will be planted in the field at Aberdeen in the spring of 2000 for seed
increase.
Evaluation of Semidwarf
Barleys from the Barley Genetic Stock Collection
An Hang, J.D. Franckowiak and K.
Satterfield; USDA-ARS, Aberdeen, ID., North Dakota State University, Fargo, ND,
and University of Idaho, Aberdeen, ID, USA
The
seven lines used in this study, were among 565 barley genetic stocks obtained
from Dr. J.D. Franckowiak, North Dakota State University (Franckowiak and
Pecio, 1991). These specific lines were derived from crosses between Bowman, a
two-rowed, drought tolerant cultivar and various mutant stocks. The genetic
stocks, many of which were semidwarf types, were planted in 1996 at Aberdeen,
ID for seed increase. Thirty-four semidwarf lines were selected and planted
under irrigation at Aberdeen in 1997 to compare their agronomic performance
with the normal height parent Bowman. After screening these semidwarf lines,
seven were selected to evaluate in the field for the 1998 and 1999 growing
seasons at two Idaho locations, under irrigation at Aberdeen and on dryland at
Tetonia. These selected lines were planted along with the cultivar Bowman in
four-row, 2.4 meter long plots with four replications in a randomized complete
block design. Only the two center rows were harvested. Heading date, plant
height, seed weight, and seed yield were recorded.
At
both locations, the average heading dates of the semidwarf derived lines were
similar to Bowman, with the exception of a line derived from a ‘Triumph’ and
Bowman cross. This line tended to head 5-6 days later at both locations. The
average plant height of the semidwarf lines was 3.8 cm to 20.9 cm shorter than
Bowman on dryland, with the exception of the line Bowman*5/DWS 1249 HJA 80089,
which nearly equaled Bowman under dryland conditions. Under irrigation at
Aberdeen, the average plant height of the semidwarf lines was 4.5 cm to 22.9 cm
shorter than Bowman. Seed weight and seed yield of derived lines were very
comparable to Bowman under irrigation at Aberdeen, although one line, Triump/4*
Bowman, yielded higher than Bowman (Table 1). On dryland at Tetonia, none of
the semidwarf derived lines equaled Bowman in seed weight and seed yield (Table
2). Our study indicated that under irrigated conditions, semidwarf derived
lines performed as well as the normal height parent and bettered the parent in
one case. However, on dryland these lines cannot compete with the normal height
parent. As mentioned above, Bowman is a drought tolerant cultivar and well
adapted in many non-irrigated areas. The semidwarf derived lines used in our
study were from different sources including Jotun mutant, Triumph, erectoides
genes, and others. Semidwarf lines derived from Jotun and ‘Diamond’ have been
frequently selected in barley breeding programs (Pecio and Franckowiak, 1989).
Previous studies showed that semidwarf genes in barley are often associated
with many undesirable characteristics such as late maturity, lower yield, lower
test weight, lower kernel plumpness, and disease susceptibility (Ali et al., 1978; Foster and Thompson, 1987;
Thomas et al., 1991; Nedel et al., 1993; Hellewell and Rasmusson,
1995). Our results indicate that environment is an important limiting factor
for the deployment of the semidwarf genes. The average annual rainfall at
Tetonia is approximately 44 cm. Under such condition, breeding for dwarf or
semidwarf barley lines seems to be unnecessary because the crop naturally tends
to be short on dryland. Development of semidwarf barleys for irrigated
environments in Idaho and the Western United States is more practical and can
be achieved.
Table 1: Means
of Three Years Agronomic Data for Lines Derived from Bowman Crossed with
Semidwarf Barleys under Irrigation at Aberdeen, ID.
|
Line |
Heading Day |
Height (cm) |
100-Seed Weight (gm) |
Seed Yield |
|
Bowman |
196 |
97.4 |
4.9 |
1100 |
|
Bowman*7/DWS1234
Jotun Derv. |
197 |
74.5 |
4.8 |
1177 |
|
Bowman*5/DWS1249
HJA 80089 |
196 |
90.8 |
5.1 |
1066 |
|
Bowman*4DWS1072SA6102
2-2-4-8sdw |
196 |
92.9 |
5.2 |
1048 |
|
DWS
1048OUM168/6*Bowman |
197 |
77.1 |
4.0 |
1065 |
|
DWS
1121/ert-zc.149/3*Bowman |
197 |
78.1 |
4.6 |
1073 |
|
Mat-f.23/2*Bowman |
196 |
88.1 |
4.9 |
1094 |
|
Triumph/4*Bowman |
201 |
80.3 |
4.8 |
1302 |
Table 2: Means of Three Years Agronomic Data for Lines
Derived from Bowman Crossed with Semidwarf Barleys on Dryland at Tetonia, ID.
|
Line |
Heading Day |
Height (cm) |
100-Seed Weight (gm) |
Seed Yield |
|
Bowman |
196 |
73.7 |
4.6 |
1006 |
|
Bowman*7/DWS1234 Jotun Derv |
197 |
53.4 |
4.0 |
826 |
|
Bowman*5/DWS1249 HJA 80089 |
196 |
74.1 |
4.5 |
945 |
|
Bowman*4DWS1072SA6102 2-2-4-8sdw |
196 |
69.6 |
4.4 |
795 |
|
DWS 1048OUM168/6*Bowman |
197 |
60.0 |
3.7 |
820 |
|
DWS 1121/ert-zc.149/3*Bowman |
197 |
60.0 |
4.3 |
865 |
|
Mat-f.23/2*Bowman |
196 |
65.8 |
4.2 |
873 |
|
Triumph/4*Bowman |
201 |
58.4 |
4.1 |
882 |
References
Ali,
M.A.M., S.O. Okiror and D.C. Rasmusson. 1978. Performance of semidwarf barley.
Crop Sci. 18:418-422.
Foster,
A.E. and A.P. Thompson. 1987. Effect of a semidwarf gene from Jotun on
agronomic and quality traits of barley. pp. 979-982. In S. Yasuda and T. Konishi (eds) Barley Genentics V. Proc. Fifth
Intl. Barley Genet. Symp., Okayama 1986. Sanyo Press Co., Okayama.
Franckowiak,
J.D. and A. Pecio. 1991. Coordinator’s report: Semidwarf genes: A listing of
genetic stocks. Barley Genetics Newsletter 21:116-125.
Hellewell,
K.B. and D.C. Rasmusson. 1995. Yield enhancement of semidwarf barley. Agron.
Abstr. p. 83.
Nedel,
J.L., S.E. Ullrich, J.A. Clancy and W.L. Pan. 1993. Barley semidwarf and
standard isotype yield and malting quality response to nitrogen. Crop Sci.
33:258-263.
Pecio,
A. and J.D. Franckowiak. 1989. Agronomic effects of semidwarf gene in barley.
Barley Newsletter 33:101.
Thomas,
W.T.B., W. Powell and J.S. Swanston. 1991. The effects of major genes on
quantitatively varying characters in barley. 4. The G pert and denso loci and
quality characters. Heredity 66:381-389.
Coordinator's report:
Trisomic and aneuploid stocks
A. Hang
USDA-ARS, National Small Grains Germplasm
Research Facility,
Aberdeen, Idaho 83210, USA
Seven
primary trisomics of cultivar SE16 were planted in the greenhouse for seed
increase.
A
compensating diploid with 2n=12 + 2 acros 6HSL + 2 acros 6HLS
derived from a cross between a primary trisomic for chromosome 6H and chlorina
seedling 13 (fch13) was used to cross
to three barley cultivars: Betzes, Bowman, and Klages. Seeds derived from
backcross F4 plants will be planted at two locations in Idaho for
evaluating agronomic characteristics.
Reference
Hang,
A., C.S. Burton and K. Satterfield. 1998. Nearly compensating diploids
involving chromosome 6(6H) in barley (Hordeum
vulgare L.). J. Genet. and Breed. 52:161-165.
Aneuploid analysis of yst3 mutant
A. Hang and K. Satterfield, USDA-ARS and
University of Idaho, Aberdeen, ID
Yellow
streak 3 (yst3) mutant has been
described by Tsuchiya, 1984 and Franckowiak, 1997. Primary and telotrisomic
analyses have been used unsuccessfully to map the gene (Wang and Tsuchiya,
1990). Wang, 1992 and Tsuchiya and Alanko, 1976 associated yst3 to chromosome 2H, even though data analyses from telotrisomic
2HL and telotrisomic 2HS (Wang, 1992) did not support the conclusion. Wang,
1992 assumed that the telotrisomic for the chromosome 2HS he used might have
had an abnormality in the chromosomal structure. We crossed yst3 to primary trisomic 5H,
telotrisomic 2HL and telotrisomic 2HS. F2 segregation data are
presented in Table 1. The results showed that high numbers of yst3 mutants were found in the trisomic
portion of the F2 population from crosses between telotrisomic 2HL
and 2HS and yst3, indicating that
this gene is not associated with 2HL or 2HS. This finding corresponds with
Wang’s data, 1992. In a F2 population of 230 crosses between triplo
5H and yst3, we found no recessive
mutant in the trisomic portion. This indicates that yst3 might be associated with chromosome 5H.
Table
1. Segregation ratios in F2 of crosses between trisomic types and
yellow streak 3 (yst3).
|
|
2x |
2x
+ 1 |
Total |
|
||||||
|
|
+ |
yst3 |
+ |
yst3 |
+ |
yst3 |
Total |
|||
|
|
|
|
|
|
|
|
|
|||
|
Triplo 2HL/yst3 |
48 |
23 |
30 |
9 |
78 |
32 |
110 |
|||
|
Triplo 2HS/yst3 |
68 |
20 |
47 |
13 |
115 |
33 |
148 |
|||
|
Triplo 5H/yst3 |
135 |
25 |
70 |
0 |
205 |
25 |
230 |
|||
Based
on linkage drag from Bowman backcross data, Franckowiak, 1995 mapped this gene
on chromosome 3HS. But trisomic analysis in the F2 population with
chromosome 3H showed that among 30 plants from the trisomic portion, 7
recessive mutants (yst3) were
observed (Tsuchiya and Alanko, 1976). This means yst3 is not located on chromosome 3. Because of the controversial location of this gene and because
the expression of this gene is affected by environment, especially by
temperature (Franckowiak, 1997), the yst3
is not a good marker to use.
References:
Franckowiak,
J.D. 1995. Note on linkage drag in
Bowman backcross derived lines of spring barley. Barley Genetics Newsletter
24:63-70.
Franckowiak,
J.D. 1997. BGS 462, Yellow streak 3 (yst3).
Barley Genetics Newsletter 26:409.
Tsuchiya,
T. and J.V. Alanko. 1976. Genetic studies of two mutations by means of primary
trisomic analysis. Barley Genetics Newsletter 6:84-85.
Tsuchiya, T. 1984. BGS 462, yellow streak 3 (yst3). Barley Genetics Newsletter 14:99.
Wang, S.
and T. Tsuchiya. 1990. Primary trisomic and telotrisomic analyses of yst3 in barley. Barley Genetics
Newsletter 19:62-63.
Wang, S.
1992. Primary trisomic analysis of the yellow stripe 3 (yst3) gene in barley. Barley Genetics Newsletter 21:73-74.
Coordinator's report:
Translocations and balanced tertiary trisomics
Gottfried Künzel
Institute of Plant Genetics and Crop
Plant Research,
DE-06466 Gatersleben, Germany
On
the basis of 240 translocation breakpoints (TBs) of 120 T lines,
cytogenetically integrated physical RFLP maps of high resolution were
constructed for the seven barley chromosomes. Simultaneously, the
karyologically definable TB positions could be remarkably refined by the use of
a software program specifically designed for that purpose (Künzel et al., 2000). At present, these 120 T
lines, most precisely defined as to their TB positions, might represent a
special kind of 'Core Collection'. Images of the physically integrated maps
together with tables indicating the map positions of the 240 TBs and idiograms
of the 120 T-lines are available from GrainGenes at ”http://wheat.pw.usda.gov/ggpages/Barley_physical/”
and
”http://wheat.pw.usda.gov/ggpages/Barley_physical/Idiograms/”, respectively.
There is no new
information on balanced tertiary trisomics since the latest report in BGN 23.
For seed requests please ask the coordinator.
Reference
Künzel, G., Larissa
Korzun and A. Meister. 2000. Cytologically integrated physical RFLP maps for
the barley genome based on translocation breakpoints. Genetics: 154 (in press).
Coordinator's report:
Autotetraploids
Wolfgang Friedt
Institute of Crop Science and Plant
Breeding I
Justus Liebig-University
Ludwigstrasse 23, DE-35390 Giessen,
Germany
E-mail:
wolfgang.friedt@agrar.uni-giessen.de
Fax: +49 641 99 37429
There is no new information about Autotreploids. A complete list of available barley autotetraploids is descriebed in former issues of Barley Genetics Newsletter (cf. BGN22:103-109 and BGN 23:164-172). These stock lists are still valid and up-to-date. The stocks are maintained at the Institute of Crop Science and Plant Breeding I in Giessen, Germany, and seed requests can be made to the Coordinator for Autotetraploids at any time. Only small samples can be delivered.
Coordinator's report:
Disease and pest resistance genes
Brian J. Steffenson
Department of Plant Pathology
North Dakota State University
Fargo, North Dakota 58105, USA
The genetics of powdery mildew resistance was studied in the
doubled haploid population Harrington/TR306 to several isolates of Blumeria (=Erysiphe) graminis f. sp.
hordei (Falak et al., 1999). Based on observed seedling infection types, the
population was classified into four groups. The number of progeny in these
groups fit a phenotypic ratio for a two-gene model. One of the two resistance
loci mapped to the same location as Mlg
(contributed by Harrington), which was previously mapped to chromosome 4 (4H)
in the Harrington/TR306 population by Steffenson and Kleinhofs, 1995. The other
resistance gene mapped to chromosome 7 (5H) and was tentatively designated Ml(TR)
because it was contributed by parent TR306. Evaluation of the progeny in the
field revealed, in addition to Mlg and
Ml(TR), two QTL contributing to resistance on chromosomes 6 (6H) and 7
(5H).
Qi et al., 1999
investigated QTL for partial resistance (as measured by latent period) to Puccinia hordei (the leaf rust pathogen;
isolates 1.2.1 and 24) in a recombinant inbred line population of the cross
Vada/L94 at both the seedling and adult plant growth stages. Three and five QTL
were identified at the seedling and adult plant stages, respectively, to
isolate 1.2.1 in a previous study (Qi et
al., 1998). In a later study (Qi et
al., 1999), one additional seedling resistance QTL and three additional
adult plant resistance QTL were identified in Vada to isolate 24. These were
designated Rphq7, Rphq8, Rphq9, and Rphq10 and
mapped to chromosomes 7 (5H), 1 (7H), 1 (7H), and 4 (4H), respectively. As in
the 1998 study, Qi et al., 1999
identified resistance QTL that were growth stage dependent: Rphq7 was only effective at the seedling
stage, whereas Rphq8, Rphq9, and Rphq10 were only effective at the adult plant stage. Overall, ten
QTL were identified for partial resistance in Vada to the two isolates of the
pathogen, 1.2.1 and 24 (Qi et al.,
1998, 1999). Significantly, several resistance QTL were isolate specific. Rphq7, Rphq8, Rphq9, and Rphq10 were only effective against
isolate 24, whereas Rphq5 and Rphq6 were only effective against
isolate 1.2.1. Only two QTL (Rphq2 and
Rphq3) were effective at both growth stages and appeared to be isolate
non-specific.
The rym3 gene
controlling reaction to Barley Yellow Mosaic Virus (BaYMV) was first described
in Ea 52, a barley mutant induced from the BaYMV susceptible cultivar Chikurin
Ibaraki 1 by gamma ray irradiation (Ukai and Yamashita 1980). To determine the
chromosomal position of this gene, Saeki et
al., 1999 constructed a partial molecular marker map of the cross Ishuku
Shirazu (carrying rym3)/Ko A (BaYMV
susceptible) and evaluated the progeny for reaction to BaYMV. The rym3 gene was mapped to chromosome 7
(5H) and is flanked by the RFLP markers ABG705A (11.7 cM distal) and MWG28 (7.2
cM proximal).
Konishi (2000; see this volume) proposed a new nomenclature system
for genes conferring resistance to BaYMV and Barley Mild Mosaic Virus (BaMMV).
Konishi recommends that genes controlling reaction to BaYMV be designated rym (reaction to barley yellow
mosaic virus) and those controlling reaction to BaMMV be designated rmm (reaction to barley mild
mosaic virus). Although the two viruses are similar in a number of
respects and are both members of the bymovirus group, they differ in several
important properties (e.g. serology and RNA sequences), most notably their
reaction on barley genotypes. Konishi recommends this gene symbol change to
avoid confusion concerning the specific virus resistance conferred by these
genes.
Simón 1997 studied partial resistance to leaf rust in 116-5, a
line originally derived from the cross L94/Cebada Capa. The number of loci
contributing to partial resistance as measured by latent period was estimated
at 7 to 8. An association was found between the vrs1 (v) locus
(controlling the six-rowed spike character on chromosome 2 (2H)) and partial
resistance to leaf rust, suggesting that genes contributing to latent period
reside on chromosome 2 (2H).
Williams et al., 1999
studied resistance to Pyrenophora teres
f. maculata (the spot form of net
blotch pathogen) in the doubled haploid population Galleon/Haruna Nijo.
Seedlings were evaluated to a mixture of five isolates of P. t. f. maculata, and when a reaction score of 4
(based on a 1 to 9 scale) was used as a threshold for classifying resistant and
susceptible plants, a 1:1 segregation was found for the 95 DH lines in the
population. This indicates that a single gene (proposed locus symbol of Rpt4; resistance to Pyrenophora teres) confers
resistance to P. t. f. maculata in
cultivar Galleon. Rpt4 confers
dominant resistance and is apparently effective at the adult plant stage. QTL
analysis was performed to identify the chromosomal position of the resistance
locus. A QTL of major effect (explaining 80% of the reaction variation) was
positioned on chromosome 1 (7H) flanked by the RFLP markers CDO673 and PSR117.
A review of the nomenclature for this and other genes controlling reaction to P.
teres is forthcoming.
Resistance gene cloning and mapping of
resistance gene homologues and pathogen related genes.
Using
tightly linked genetic markers, Wei et al.,
1999 developed a physical contig of YAC and BAC clones that spanned the powdery
mildew resistance gene cluster at the locus
Mla. The Mla resistance
cluster was associated with nucleotide binding site-leucine rich repeat
(NBS-LRR) gene families within a 240 kb interval on chromosome 5 (1H).
Leister
et al., 1999 mapped resistance gene
analogs (of the NBS-LRR type) in barley and found linkage with the Mla locus for powdery mildew resistance
and the Rpg1 locus for stem rust
resistance. In a study to elucidate the interaction of barley with the stripe
fungus Pyrenophora graminea,
Pecchioni et al., 1999 mapped
pathogen related (PR) genes. Tha (thaumatin-like
locus) and Chi1 (chitinase 1) were
mapped to chromosome 1 (7H), Prx7
(peroxidase 7) was mapped to chromosome 2 (2H), Glb32 (b-(1-3)-glucanase
isoform 32) to chromosome 3 (3H), Ftt (a fourteen three three
(14-3-3) protein analog produced in response to powdery mildew infection) to
chromosome 4 (4H), Chs3 (chalcone
synthase) to chromosome 5 (1H), and Rip1
(ribosome inactivating protein 1) to chromosome 7 (5H).
Assigning new locus and
allele symbols for disease and pest resistance genes.
To
assign a new locus and allele symbol for disease and pest resistance genes in
barley, it is incumbent upon the investigator(s) to provide evidence that 1)
the resistance is conferred by a single gene, 2) the gene confers a unique
infection response or reaction pattern compared to other ”known” genes, and 3)
allelism tests with potential alleles are negative and/or the gene maps to a
unique location. These criteria are based on those recommended by Franckowiak et al., 1997 for leaf rust resistance
genes, but are applicable for other resistance genes. In assigning locus
symbols, it is recommended that the simple and sensible rules proposed by
Moseman 1972 be used. To validate a new locus and allele, please send the
appropriate information to me prior to publication, and I will post it on the
Graingenes news group for all interested researchers to review. The proposed
locus and allele will become validated if other researchers make no objections.
It
is desirable to deposit both the original source of the resistance gene (i.e. a
pure seed increase from a single plant selection) and the isolate of the
pathogen used to identify the gene in an international germplasm and culture
repository, respectively. This would ensure that these valuable materials are
preserved indefinitely. The accession number and repository location could then
be included in the publication validating the new locus and allele.
References
Falak, I., D.E. Falk, N.A. Tinker and D.E. Mather. 1999.
Resistance to powdery mildew in a doubled haploid population and its
association with marker loci. Euphytica 107:185-192.
Franckowiak, J.D., Y. Jin and B.J. Steffenson. 1997. Recommended
allele symbols for leaf rust resistance genes in barley. Barley Genetics
Newsletter 27:36-44.
Konishi, T. 2000. Proposed gene symbols for resistance to Barley
Mild Mosaic Virus (BaMMV) in barley. Barley Genetics Newsletter 30: (this
volume).
Leister, D., J. Kurth, D.A. Laurie, M. Yano, T. Sasaki, A. Graner
and P. Schulze-Lefert. 1999. RFLP- and physical mapping of resistance gene
homologues in rice (O. sativa) and barley (H. vulgare). Theor. Appl.
Genet. 98:509-520.
Moseman, J.G. 1972. Report on genes for resistance to pests.
Barley Genetics Newsletter 2:145-46.
Pecchioni, N., G. Valè, H. Toubia-Rahme, P. Faccioli, V. Terzi and
G. Delogu. 1999. Barley-Pyrenophora
graminea interaction: QTL analysis and gene mapping. Plant Breed.
118:29-35.
Qi, X., G. Jiang, W. Chen, R.E. Niks, P. Stam and P. Lindhout.
1999. Isolate-specific QTLs for partial resistance to Puccinia hordei in barley. Theor. Appl. Genet. 99:877-884.
Qi, X., R.E. Niks, P. Stam and P. Lindhout. 1998. Isolate-specific
QTLs for partial resistance to Puccinia
hordei in barley. Theor. Appl. Genet. 96:1205-1215.
Saeki, K, C. Miyazaki, N. Hirota, A. Saito, K. Ito and T. Konishi.
1999. RFLP mapping of BaYMV resistance gene rym3
in barley (Hordeum vulgare). Theor.
Appl. Genet. 99:727-732.
Simón, M.R. 1997. Asociaciones entre marcadores morfológicos y
bioquímicos con la resistancia a roya de la hoja de cebada (Puccinia hordei Otth). Agriscientia
14:3-9.
Steffenson,
B.J. and A. Kleinhofs. 1995. Genetics and mapping of powdery mildew resistance
in the Harrington/TR306 and Harrington/Morex doubled haploid populations.
Barley Newsletter 39:89-91.
Ukai, Y. and A. Yamashita. 1980. Induced mutation for resistance
to barley yellow mosaic virus. Jpn. J. Breed. 30:125-130.
Wei, F., K. Gobelman-Werner, S.M. Morroll, J. Kurth, L. Mao, R.
Wing, D. Leister, P. Schulze-Lefert and R.P. Wise. 1999. The Mla (powdery mildew) resistance cluster
is associated with three NBS-LRR gene families and suppressed recombination
within a 240-kb DNA interval on chromosome 5S (1HS) of barley. Genetics
153:1929-1948.
Williams, K.J., A. Lichon, P. Gianquitto, J.M. Kretschmer, A.
Karakousis, S. Manning, P. Langridge and H. Wallwork. 1999. Identification and mapping of a gene conferring
resistance to the spot form of net blotch (Pyrenophora
teres f. maculata) in barley.
Theor. Appl. Genet. 99:323-327.
Coordinator's report: Eceriferum Genes
Udda Lundqvist
Svalöf Weibull AB
SE-268 81 Svalöv, Sweden
e-mail:udda@ngb.se
No
new research work on eceriferum genes
has been reported since the latest reports in Barley Genetics Newsletter (BGN).
All descriptions made in the special volume of Barley Genetics Newsletter (BGN
26) are still up-to-date and valid. The publication of these descriptions is
available under the address:
http://wheat.pw.usda.gov/ggpages/bgn/
Notice
that the address is changed since the latest issue of Barley Genetics
Newsletter, BGN 28:
Current
lists of new and revised BGS descriptions will be presented in each volume of
BGN.
The
incorporation and conversion of the descriptions of the eceriferum loci into the International Triticeae Genome Database
”GrainGenes” according to the ACEDB format is still in progress and will
hopefully be fulfilled before the next International Barley Genetics Symposium
in October 2000. Some major problems have to be solved in order to make the
Database successfull. Also the feeding of the images of the different eceriferum genes is still in progress.
More pictures have been taken during the summer season of 1999 in Aberdeen, ID,
USA.
Many
of the stocks of these genes are available at the Genetics Stock Center in
Aberdeen, ID, USA. The Swedish eceriferum
loci with all the valuable alleles are available at the Nordic Gene Bank,
Box P.O. 41, SE-230 53 Alnarp, Sweden.
FAX:
+46 40 536650, e-mail: nordgen@ngb.se
Researchers
in the field of eceriferum genes, ”Glossy sheath” and ”Glossy leaf” are
urgently requested to report matters of interest and research to the
coordinator as well. Seed requests can be forwarded to the Nordic Gene Bank
regarding the Swedish mutants, all the others to the Genetics Stock Center or
to the coordinator at any time.
Coordinator's
report: Nuclear genes affecting the chloroplast
Diter von Wettstein
Department of Crop and Soil Sciences,
Washington State University
Pullman WA 99164-6420, USA
E-mail: diter@wsu.edu
The stock list and genetic information
presented in the Barley Genetics Newsletter 21: 102-108 is valid and
up-to-date. The stocks have been transferred to the Nordic Gene Bank. Requests
for stocks available for distribution are to be sent to:
Dr.
Mats Hansson
Department
of Biochemistry
Center
for Chemistry and Chemical Engineering
Lund
University
P.O.Box
124
SE-221
00 Lund SWEDEN
Phone:
+46-46-222 0105
Fax: +46-46-222 4534
E-mail:
Mats.Hansson@biokem.lu.se
Recent references
Hansson
A., C.G. Kannangara, D. von Wettstein and M. Hansson M. 1999. Molecular basis
for semidominance of missense mutations in the XANTHA-H (42-kDa) subunit of
magnesium chelatase. Proc.Natl. Acad. Sci.USA 96: 1744-1749.
Wettstein
D., von. 2000. Chlorophyll Biosynthesis I: From analysis of mutants to genetic
engineering of the pathway. Chapter in Discoveries in Plant Biology. S.D. Kung
& S.F.Yang (eds.) Vol.3 p. 1-19 (in press).
Wettstein
D., von. 2000. Chlorophyll Biosynthesis II: Adventures with native and
recombinant enzymes. Chapter in Discoveries in Plant Biology. S.D. Kung &
S.F. Yang (eds.) Vol. 3 p. 1-48 (in press).
Coordinator's report: The
Genetic Male
Sterile Barley Collection
M.C. Therrien
Agriculture and Agrifood Canada
Brandon Research Centre
Box 1000A, RR#3, Brandon
Manitoba, Canada R7A 5Y3
E-mail: Mtherrien@em.agr.ca
The
GMSBC has been at Brandon since 1992. If there are any new sources of
male-sterile genes that you are aware of, please advise me, as this would be a
good time to add any new source to the collection. For a list of the entries in
the collection, simply E-mail me at the above address. I can send the
file (14 Mb) in Excel format. We continue to store the collection at -20°C and
will have small (5 g) samples available for the asking. Since I have not
received any reports or requests in 1999, there is no summary in my report.
Coordinator's
Report: Inversions
B.O.Bengtsson
Department of Genetics, University of
Lund
Sölvegatan 29, SE-223 62 Lund, Sweden
E-mail: bengt-olle.bengtsson@gen.lu.se
There
is no new information on stocks of barley inversions. Every contribution and
other research reports in this field are welcome to be sent to the coordinator.
Coordinator's report: Ear Morphology
Genes
Udda Lundqvist
Svalöf Weibull AB
SE-268 81 Svalöv, Sweden
Not
much new research work on the different ear morphology genes has been reported
since the latest reports in Barley Genetics Newsletter (BGN). All descriptions
made in the special volume of Barley Genetics Newsletter (BGN 26) and later
issues are still up-to-date and valid. The publications of these descriptions
are available under the address:
http://wheat.pw.usda.gov/ggpages/bgn/
Notice
that the address is changed since the latest issue of Barley Genetics
Newsletter, BGN 28.
Current
lists of new and revised BGS descriptions will be presented in each volume of
BGN.
The
incorporation and conversion of the descriptions of the Ear Morphology Genes
into the International Triticeae Genome Database ”GrainGenes” according to the
ACEDB format is still in progress and will hopefully be fulfilled before the
next International Barley Genetics Symposium in October 2000. Some major
problems have to be solved in order to make the Database successful. Also the
feeding of the images of the different Ear Morphology Genes is still in
progress. Another 1000 pictures have been taken during the summer season of
1999 in Aberdeen, ID, USA.
Many
of the stocks of these genes are available at the Genetics Stock Center in
Aberdeen, ID, USA. The Swedish ”Ear Morphology” loci with all their many
valuable alleles are available at the Nordic Gene Bank, Box P.O.41, SE-230 53
Alnarp, Sweden.
FAX:
+46 40 536650, e-mail: nordgen@ngb.se
Researchers
in the field of ear morphology genes are urgently encouraged to submit matters
of interest and research to the coordinator as well. Seed requests can be
forwarded to the Nordic Gene Bank regarding the Swedish material, all the
others to the Genetics Stock Center or to the coordinator at any time.
Coordinator's
report: Semidwarf genes
J.D. Franckowiak
Department of Plant SciencesNorth Dakota
State University
Fargo, ND 58105, U.S.A.
Börner et al., 1999 mapped the position of two recessive dwarfing mutants
in chromosome 2H. The gibberellic acid (GA) insensitive dwarf gai (Hv287), which co-segregated with
RFLP markers MWG2058 and MWG2287, is in chromosome 2HS close to the centromere.
The GA sensitive mutant gal (Hv288),
which co-segregated with RFLP markers MWG581 and MWG882A, is in chromosome 2HL
about 22 cM distal from the six-rowed spike 1 (vrs1) locus and 55 cM from the gai
locus. Ivandic et al., 1999 mapped a
dominant GA insensitive dwarf mutant (Dwf2,
93/B694) in chromosome 4HS. They suggested that the Dwf2 mutant, which shows an extremely prostrate growth habit, may
be homoeologous with GA insensitive mutants Rht3
and Rht10 of wheat.
Identification of new semidwarf mutants
that can be used to improve barley has been rather unsuccessful because most
induced mutants are associated with reduced plant vigor and lower yields. Only
mutants at the semidwarf 1 (sdw1)
locus in chromosome 3HL are widely used to reduce plant height in barley
cultivars. Bregitzer et al., 1998 and
Molina-Cano et al., 1999 reported recently that mutants induced in semidwarf
cultivars are inferior to their parents. Bregitzer et al., 1998 found that somaclonal variants isolated from the
cultivar Golden Promise were shorter and lower yielding than their parent.
Molina-Cano et al., 1999 found that
an ABA-insensitive mutant (TL43) for reduced dormancy in the cultivar Triumph
also reduced plant height and grain yield when tested in Spain. Both Golden
Promise and Triumph have genes for reduced plant height, ari-e.GP at the breviaristatum-e locus and sdw1.d at the semidwarf 1 locus, respectively. Yin et al., 1999 examined recombinant inbred
lines from a Prisma by Apex cross and found that most traits controlling plant
height, yield, and yield-determining physiological traits were associated with
the sdw1.d or denso allele from Prisma. These reports suggest that plant height
mutants in barley often increase the sensitivity of barley to environmental
stresses.
Semidwarf
genes in Midwestern USA barley cultivars.
Control of plant
height can reduce lodging and increase yield and yield stability in barley;
however, major genes that control culm length have not been identified in
six-rowed barley cultivars adapted to the Upper Midwest of the USA. Mutants at
the sdw1 locus provide acceptable
height control in some barley production areas, but sdw1 mutants have not been successfully incorporated into material
adapted to the Upper Midwest, a semi-arid environment where heat and drought
stress frequently occur. Most cultivars grown in this area have a six-rowed
spike type and are derived from crosses to introductions from Northeastern
China.
Peduncle
length is positively associated with yield.
If semidwarf
mutants tend to reduce productivity in barley, other genes that stabilize
performance must also exist. Two recent reports indicate that peduncle length
is positively associated with yields and yield stability in six-rowed
cultivars. Fufa and Gebre, 1998 examined 17 cultivars in Ethiopia under
residual moisture conditions and Yahyauoui et
al., 1998 studied a collection of 296 landraces in Tunisia. Among spring
barley cultivars bred for the USA, the long peduncle trait is frequently
observed in six-rowed cultivars. Long peduncles are also a characteristic of wild
barley (Hordeum vulgare subsp. spontaneum) accessions. These
observations indicate that peduncle length may provide a visual assessment of
drought and heat stress tolerance in barley.
Development
of the cultivar Bowman from two- by six-rowed crosses. Many
researchers have crossed two- and six-rowed cultivars to improve barley and
have reported that F1 plants are much taller than either parent
(Smith, 1951). Tedin and Tedin, 1926 reported that six-rowed segregates were
shorter than two-rowed or intermediate segregates. The gene symbol h (now hcm for short culm) was assigned. Neatby, 1929 demonstrated this
locus is linked to the v (now vrs1 for six-rowed spike 1) locus in
chromosome 2HL. Subsequent studies found that plant height has a fairly high
broad sense heritability and can be associated with photoperiod responses (see
Hockett and Nilan, 1985). A complex series of crosses between two- and
six-rowed cultivars were used to develop the cultivar Bowman for western North
Dakota (Franckowiak et al., 1985).
Bowman has height and maturity characteristics similar to those in its
six-rowed parents. Other two-rowed lines with a similar growth pattern have
been recovered only when Bowman is used as a parent.
Plant
height control in Bowman and Bowman backcross-derived lines. Because
Bowman does not produce extremely tall F1 plants when crossed to
Midwestern six-rowed cultivars as Morex, it is believed to carry a Vrs1.b - hcm1.a recombinant. Crosses between Bowman and two-rowed cultivars
also do not produce extremely tall plants; thus, Bowman likely has some height
control genes from its two-rowed parents also. Bowman has been used as the
recurrent parent in the development of backcross-derived lines for many
morphological markers. However, none of the derived lines seems suitable for
study of the linkage distance between the vrs1
and hcm loci. Hence, expression of hcm1.a gene may not be as strong in
Bowman as in Midwestern six-rowed cultivars.
Short-stature
in two-rowed barley cultivars from Mexico.
Two-rowed selections
with short stature and normal vigor were found last year in progeny from
crosses to the cultivars Shyri and Aleli, developed by the ICARDA/CIMMYT barley
breeding program in Mexico. Complex crosses between two- and six-rowed lines
are made frequently in the barley improvement program directed by Dr. Hugo E.
Vivar. Most short segregates had the Vrs1.b
gene from Bowman derived parents instead of the Vrs1.t (deficiens) allele from Shyri and Aleli. The short-stature
segregates also exhibited the dominant allele (Int-c.a) at the intermedium spike-c locus. Although its expression
is suppressed in Shyri and Aleli, the Int-c.a
gene originated from these cultivars. The short-stature segregates appeared
shorter than Bowman and had semicompact spikes. This information suggests that
Shyri and Aleli have a gene for short height linked closely to the int-c locus in chromosome 4HS and that
the short-stature gene interacts with the hcm1.a
gene from the Bowman derivatives.
Hordeum intermedium
and Bowman backcross-derived lines. If the Int-c.a allele is linked to a gene for short stature, its effects
should have been reported in previous studies of crosses between two- and
six-rowed cultivars. Cultivars with spikes similar to those of the
short-stature segregates were classified
previously as H. intermedium
(Harlan, 1918). Similar segregates were also recovered by Woodward, 1947 from
crosses between two-rowed cultivars and H.
deficiens accessions. Shakudo and Kawase in 1951 (see Nilan, 1964) reported
that two inhibitors, factors for fertility of lateral florets, reduced culm
length in barley. One inhibitor in six-rowed barley is likely the hcm1.a allele in chromosome 2HL and the
other is likely associated with the Int-c.a
allele. Since backcrossing of two mutants at the int-c locus, int‑c.5
in Bonus and vrs5.n in Gamma 4, into
Bowman did not produce short plants or dense spikes (Lundqvist and Franckowiak,
1997), mutants at the int-c locus
probably do not have a pleiotropic effect on plant height or spike density.
Thus, many six-rowed cultivars probably have a plant height gene closely linked
to Int-c.a allele.
Unfortunately, no other morphological
markers in barley have been mapped close to the int-c locus. Among the unmapped markers, those that exhibit linkage
drag with the Int-c.a allele in
Bowman backcross-derived lines might be associated with the plant height gene
in question. Four unmapped mutants (brh.m,
brachytic; sld.e, slender dwarf; sld.f; and Zeo.h, zeocriton) were associated with the large lateral trait
(Franckowiak, 1995). But only the Zeo.h
backcross-derived line is similar in plant height and spike density to the
short-stature lines selected from the Shyri and Aleli crosses. The donor parent
for the Zeo.h line was the Mo1
mutant, isolated by S.E. Ullrich, Washington State University, from the
six-rowed cultivar Morex. Spikes of Zeo.h
plants are semicompact, but they do not have the large lateral spikelets
associated with the Int‑c.a
allele. Thus, the Zeo.h line may have
the short-stature gene present in Midwest six-rowed barley cultivars. Although
allelism tests have not been conducted to verify this conclusion, the locus
symbol Zeo3 and the allele symbol Zeo3.h are suggested as identifying
symbols to facilitate consideration of linkage relationships.
Linkage
between Int-c.a and glossy node
loci. Glossy node, little or no surface waxes
on nodes of the culm, is a semidominant trait that is present in most
Midwestern six-rowed cultivars. This trait is found in neither in spring
two-rowed cultivars nor in two-rowed lines derived from crosses to six-rowed
cultivars. The Harrington/Morex mapping population of the NABGMP segregated for
the glossy node trait, but correct classification of individual lines proved
difficult. Preliminary data analysis places the gene for glossy node near a
gene for reduced awn length in chromosome 4HS (Kleinhofs and Franckowiak,
unpublished).
Development of a Bowman backcross-derived
line for glossy node gene has been difficult because the derived lines do not
resemble Bowman. They are more robust and have wide leaves, large lateral
spikelets, and semicompact spikes. The glossy node stock used as a donor parent
was provided by T.M. Blake from the R.F. Eslick’s collection of barley mutants
at Montana State University. Since that stock was held under the name yellow
node (Yn), the recommended locus
using a three letter code symbol would be Ynd.
These notes suggest that the glossy node trait (Ynd1.a) is linked in coupling with the Int-c.a and Zeo3.h
alleles in six-rowed cultivars.
Development
of two-rowed lines with the Zeo3.h
gene. As outlined above, at least two
interactive plant height control systems that may be present in Midwestern
six-rowed barley cultivars. Plant height genes are closely linked to the two
loci, int-c and vrs1, primary responsible for spike type in barley. The likely
locus orders are Gth hcm vrs1 in chromosome 2HL and Zeo3 Ynd Int‑c in
chromosome 4HS. Recovery of short-stature two-rowed lines from a two- by
six-rowed cross requires recombination in both regions and should be possible
if other undesirable genes are not linked to these complexes. Midwestern
six-rowed cultivars have the dominant allele (Gth1.a) at the toothed lemma locus while many European two-rowed
cultivars have the recessive allele (gth1.b).
Often two-rowed segregates with the gth1.b
allele have higher malt extract values than those with the Gth1.a allele. Many two-rowed cultivars have a recessive gene for
late maturity about 13 cM proximal from the Gth
locus and near the glossy sheath 5 (gsh5)
locus in chromosome 2HS.
The phenotypic effects of alleles at the Zeo3 locus will be difficult to quantify
because they interact with hcm
alleles and probably other plant height genes. However, data on a population in
which segregation for plant height occurred only at the Zeo3 may have been published by Zhu et al., 1999. One parent of their DH population, CMB643, was
derived a Shyri cross and has the genotype Vrs1.t Int-c.a Zeo3.h, and the other
parent, Gobernadora, has a normal two-rowed genotype Vrs1.b int-c.b zeo3. Both
parents probably have the dominant allele at the hcm locus. Zhu et al.,
1999 found that both the vrs1 and int-c loci are associated with QTLs for
Fusarium head blight (FHB) reaction and toxin accumulation. Susceptibility to
FHB and larger lateral spikelets were associated with both the Int-c.a allele from CMB643 and with the Vrs1.b allele from Gobernadora. They
also reported a strong association between the Int-c.a allele and reduced plant height.
Much more information about plant height
control schemes in barley and their interactions with photoperiod and
environmental factors is needed. For example, six-rowed barley cultivars may
contain also a recessive plant height gene(s) for the long peduncle trait. Yet,
what is known about height control genes in six-rowed barley may be adequate to
evaluate their potential for increasing the yield and yield stability of barley
in semiarid environments.
References
Börner,
A., V. Korzun, S. Malyshev and V. Ivandic. 1999. Molecular mapping of two
dwarfing genes differing in their GA response on chromosome 2H of barley.
Theor. Appl. Genet. 99:670-675.
Bregitzer,
P., S.E. Halbert and P.G. Lemaux. 1998. Somaclonal variation in progeny of
transgenic barley. Theor. Appl. Genet. 96: 421-425.
Franckowiak,
J.D., A.E. Foster, V.D. Pederson and R.E. Pyler. 1985. Registration of ‘Bowman’
barley. Crop. Sci. 25:883.
Franckowiak,
J.D. 1995. Notes on linkage drag in Bowman backcross derived lines of spring
barley. BGN 24:63-70.
Fufa,
F. and H. Gebre. 1998. Agronomic traits in barley grown under residual moisture
in Ethiopia. Rachis 17:1-2, 58-60.
Harlan,
H.V. 1918. The identification of varieties of barley. U.S. Dept. Agr. Bull. No.
622.
Hockett,
E.A. and R.A. Nilan. 1985. Genetics. p. 187-230. In D.C. Rasmusson (ed.) Barley.
Agronomy Monograph No. 26. American Society of Agronomy, Madison, WI.
Ivandic,
V., S. Malyshev, V. Korzun, A. Graner and A. Börner. 1999. Comparative mapping
of a gibberellic acid-insensitive dwarfing gene (Dwf2) on chromosome 4HS in
barley. Theor. Appl. Genet. 98:728-731.
Lundqvist,
U. and J.D. Franckowiak. 1997. BGS 176, intermedium spike-c, int-c. BGN 26:200‑201.
Molina-Cano,
J.L., A. Sopena, J.S. Swanston. A.M. Casas, M.A. Moralejo, A. Ubieto, I. Lara,
A.M. Perez-Vendrell, and I. Romagosa. 1999. A mutant induced in the malting
barley cv Triumph with reduced dormancy and ABA response. Theor. Appl. Genet.
98:347-355.
Nilan,
R.A. 1964. The cytology and genetics of barley, 1951-1962. Monogr. Suppl. 3,
Res. Stud. Vol. 32, No. 1. Washington State Univ. Press, Pullman.
Woodward,
R.W. 1947. The Ih, I, i allels in Hordeum deficiens genotypes of
barley. J. Am. Soc. Agron. 39:474-482.
Yahyauoui,
A.H., S. Rezgui and A.A. Jaradat. 1998. Barley landrace cultivars: source of
stress tolerance. p. 321-324. In Triticeae III. Proc. Third Internal. Triticeae
Symp., Aleppo, Syria, 4-8 May, 1997. Science Publishers, Inc. Enfield, USA.
Yin,
X., P. Stam, C.J. Dourleijn and M.J. Kropff. 1999. AFLP mapping of quantitative
trait loci for yield determining physiological characters in spring barley.
Theor. Appl. Genet. 99:244‑253.
Zhu,
H., L. Gilchrist, P. Hayes A. Kleinhofs, D. Kudrna, Z. Liu, L. Prom, B.
Steffenson, T. Toojinda and H. Vivar. 1999. Does function follow form?
Principal QTLs for Fusarium head blight (FHB) resistance are coincident with
QTLs for inflorescence traits and plant height in a double haploid population
of barley. Theor. Appl. Genet. 99:1221-1232.
Coordinator's report:
Anthocyanin Genes
Barbro Jende-Strid
Carlsberg Research Laboratory, Gamle
Carlsberg Vej 10, DK-2500 Valby Copenhagen, Denmark
There
is no further information on flavonoid genes or ant mutants since the last report published in Barley Genetics
Newsletter, BGN 28:103. Stock lists have been published in BGN 18:74-79, BGN
20:87-88, BGN 22:136-137 and BGN 24:162-165. Barley Genetic Stock descriptions
of the ant genes are published in BGN
29:80-99. Seed requests of the ant
stocks can be sent to the coordinator or to The Nordic Gene Bank, Box P.O.41,
SE-230 53 Alnarp, Sweden
PHONE:
+46 40 536640, FAX: +46 40 536650, e-mail: nordgen@ngb.se
Every
contribution and other research reports in this field are welcome to be sent to
the coordinator.
Coordinator's report: Early
Maturity Genes
Udda Lundqvist
Svalöf Weibull AB, SE-268 81 Svalöv,
Sweden
No
new research work on Early Maturity Genes has been reported since the latest
reports in Barley Genetics Newsletter (BGN). All descriptions made in the
special volume of Barley Genetics Newsletter (BGN 26) are still up-to-date and
valid. The publication of these descriptions is available under the address: http://wheat.pw.usda.gov/ggpages/bgn/
Notice
that the address is changed since the latest issue of Barley Genetics
Newsletter, BGN 28:
Current
lists of new and revised BGS descriptions will be presented in each volume of
BGN.
The
incorporation and conversion of the descriptions of the Early Maturity Genes
into the International Triticeae Genome Database ”GrainGenes” according to the
ACEDB format is still in progress and will hopefully be fulfilled before the
next International Barley Genetics Symposium in October 2000. Some major
problems have to be solved in order to make the Database successful. Also the
feeding of the images of the different Early Maturity Genes is still in
progress. More pictures have been taken during the summer season of 1999 in
Aberdeen, ID, USA.
Many
of the stocks of these genes are available at the Genetics Stock Center in
Aberdeen, ID, USA. The Swedish Early Maturity Genes with all the different
alleles are available at the Nordic Gene Bank, Box P.O.41, SE-230 53 Alnarp,
Sweden, FAX: +46 40 536650, e-mail: nordgen@ngb.se
Researchers
in the field of Early Maturity Genes are urgently requested to report matters
of interest and research to the coordinator as well. Seed requests can be forwarded
to the Nordic Gene Bank regarding the Swedish mutants, all the others to the
Genetics Stock Center or the coordinator at any time.
Coordinator's report:
Duplication of Chromosome Segments.
Arne Hagberg
Department of Plant Breeding Research
The Swedish University of Agricultural
Sciences
SE-268 31 Svalöv, Sweden
There
is no information on new barley duplications of chromosome segments since the
last publications in Barley Genetics Newsletter, BGN 25:114. All the chromosome
duplication stocks as reported in Barley Genetics Newsletter BGN 21:130-136 are
incorporated into the Nordic Gene Bank Their descriptions and cytological
analysis are compiled in a database kept there. The material is available to
every research worker, and seed requests of the chromosome duplication stocks
can be sent to the coordinator or to:
The
Nordic Gene Bank
Box
P.O.41
SE-230
53 Alnarp Sweden
PHONE:
+46 40 536640
FAX:
+46 40 536650
e-mail:
nordgen@ngb.se
Every
contribution and research report in this field are appreciated to be sent to
the coordinator. This is the concluding coordinator’s report on barley
duplications of chromosomal segments.
Coordinator's report:Wheat-barley genetic
stocks
A.K.M.R. Islam
Department of
Plant Science, Waite Campus,
The University
of Adelaide,
Glen Osmond,
S.A. 5064, Australia
There
is little new information to report for wheat-barley genetic stocks since the
last report. Islam reported in 1994 that two doses of barley chromosome 6H can
overcome the genetic male-sterility induced by barley chromosome arm 1HL and it
is possible to maintain chromosome 1H as a self-fertile monosomic addition line
in a disomic 6H background. Since then Islam was able to isolate a
disomic-monotelodisomic addition line carrying a pair of barley chromosome 6H and
a heteromorphic 1H/1HS pair. This new line is quite stable (94% of the progeny
possess either an entire 1H or at least the 1HS arm).
Reference:
Islam, A.K.M. Rafiqul and Kenneth W. Shepherd. 1999.
Isolation of a fertile wheat barley addition line carrying the entire barley
chromosome 1H. Euphytica
111(2):145-149.