A. Schiemann, A. Graner*, W. Friedt and F. Ordon
Institute of Crop Science and Plant Breeding I, Justus-Liebig-University,
Ludwigstr. 23, D-35390 Giessen, Germany
*Federal Centre for Breeding Research on Cultivated Plants, Institute for
Resistance Genetics, Graf-Seinsheim-Str. 23, D-85461 Grünbach, Germany
In Western Europe barley yellow mosaic disease is one of the most important diseases of winter barley today. In order to select for resistance in early generations, plants have to be mechanically inoculated with BaMMV in the greenhouse (Friedt 1983) or have to be tested in infested fields. Both procedures are time consuming and it has to be taken into account that plants showing negative ELISA-results may have escaped infection. Therefore, marker assisted selection has to be considered as a very useful tool in breeding for resistance. In this respect RFLP-markers for different resistance genes have been identified already (Graner et al. 1996) including the resistance gene ym4 (Graner & Bauer 1993). Furthermore, concerning ym4 an isozyme (Le Gouis et al. 1995) as well as different RAPD markers (Weyen et al. 1996) including the very tightly linked marker OP-Z04H660 (Ordon et al. 1995) are known. However, this primer exhibiting an additional band of about 660bp in susceptible lines shows a quite complex banding pattern especially in the region of interest (Fig. 1). Furthermore, as OP-Z04H660 is inherited in a dominant manner and linked to the resistance allele in repulsion phase it does not facilitate the identification of heterozygous susceptible plants in F2 which will segregate resistant plants in the offspring. Therefore, although OP-Z04H660 has to be considered well suited for marker assisted selection in DH lines, experiments were conducted in order to convert it in a more specific marker discriminating between homozygous and heterozygous genotypes.
Fig. 1. RAPD-pattern of OPZ-04 and the corresponding elongated primers on resistant (ym4) and susceptible barley varieties and lines (Click here)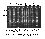
In order to achieve this as easy and as fast as possible each of the four bases - adenine(A), cytosine(C), guanine(G) and thymine(T) - were added at the 3'-end of the primer
5'-AGGCTGTGCT-3' (OP-Z04) resulting in four 11mer primers (OP-Z04A, OP-Z04C, OP-Z04G, OP-Z04T). RAPD-analysis was carried out using DNA from resistant and susceptible F1 anther derived doubled haploid (DH) barley lines out of a cross between the BaMMV/BaYMV resistant cultivar `Franka' (ym4) and the susceptible cultivar `Igri' (cf. Ordon et al. 1995). PCR reaction was performed in a volume of 25ul consisting of 25ng genomic DNA, 0.4mM dNTPs, 6mM MgCl2, 0.3uM of a random 11mer primer (Life Technologies Gibco BRL) and 1.5U AmpliTaq DNA-polymerase Stoffel-fragment (Perkin Elmer) and its corresponding reaction buffer (Perkin Elmer). The reaction mix was overlaid with mineral oil. PCR was conducted in a thermal cycler (TC480, Perkin Elmer). An intial denaturation step (94deg.C/4 min) was followed by 45 cycles of 94deg.C/1 min, 44deg.C/1 min, and 72deg.C/2 min, respectively. The heating rate from 44deg.C to 72deg.C was restricted to 5deg.C/min. Each polymerisation step was extended for 3s/cycle at 72deg.C (modified according to Sobral & Honeycutt 1993). PCR products were seperated on a 2% agarose gel (Nu Sieve, FMC) by electrophoresis and visualized by ethidium bromide staining and UV illumination (286 nm).
Out of the four 11mer primers (OP-Z04A, OP-Z04C, OP-Z04G, OP-Z04T) tested, OP-Z04A (5'-AGGCTGTGCTA-3') showed a clear cut polymorphism between resistant (ym4) and susceptible lines (Fig.1) while the other elongated primer did not exhibit any discriminating bands (data not presented). As shown in figure 1 primer OP-Z04A generates an additional 640bp band in resistant DH lines which is missing in the susceptible ones and furthermore an additional 660bp band in susceptible DH lines which is missing in the resistant ones, thereby enabling the detection of heterozygous genotypes. By testing 287 F1 anther derived DH lines out of a cross between the BaMMV/BaYMV resistant cultivar `Franka' (ym4) and the susceptible cultivar `Igri' which have been used already in studies to detect OP-Z04H660 (Ordon et al. 1995) it turned out that OP-Z04A perfectly cosegregates with OP-Z04H660. Therefore, OP-Z04A is well suited to facilitate marker-assisted selection for BaMMV-resistance encoded by ym4 in DH-lines as well as segregating F2-populations. Besides this, this primer may be well suited for high density mapping of ym4 (cf. Bauer et al. 1995). In a next step the primer OP-Z04A was elongated with additional bases and it turned out that the 12mer primer OP-Z04AT (5'-AGGCTGTGCTAT-3') and the 13mer primer OP-Z04ATA (5'-AGGCTGTGCTATA-3') exhibited the same polymorphism as OP-Z04A (Fig. 1).
In this case it is interesting to note that OP-Z04AT reveales an additional polymorphism resulting in an extra 1700bp band in resistant genotypes and two additional fragments (1500 bp and 1800 bp) in susceptible lines, cosegregating with OP-Z04H660 and OP-Z04A, respectively.
Although it turned out in this study that more specific markers may be developed very easily without sequencing, future work will now concentrate on the conversion of the RAPD marker OP-Z04A into a sequence-tagged-site (STS) marker.
Acknowledgements: Thanks are due to the Deutsche Forschungsgemeinschaft (DFG), Bonn, for financial support (Grants No. OR 72/2-1 and GR 1317/3-1).
References:
Bauer, E. and A. Graner, 1995: Basic and applied aspects of genetic analysis of the ym4 virus resistance locus in barley. Agronomie 15, 469-474.
Friedt, W., 1983: Mechanical transmission of soil-borne barey yellow mosaic virus. Phytopath. Z. 106, 16-22.
Graner, A. and E. Bauer, 1993: RFLP mapping of the ym4 virus resistance gene in barley. Theor. Appl. Genet. 86, 689-693.
Graner, A., E. Bauer, A. Kellermann, G. Proeseler, G. Wenzel and F. Ordon, 1995: RFLP-analysis of resistance to the barley yellow mosaic virus complex. Agronomie 15, 475-480.
Graner, A., E. Bauer, J. Chojecki, A. Tekauz, A. Kellermann, G. Proeseler, M. Michel, V. Valkov, G. Wenzel and F. Ordon, 1996: Molecular mapping of genes for disease resistance in barley. In: Scoles G, Rossnagel B (eds.) Proc. V Intern. Oat Conf. & VII Intern. Barley Genetics Symp. Poster Sessions Vol. 1, Saskatoon, Canada, University Extension Press, Saskatoon, Sakatchewan, pp 253-255.
Le Gouis, J., M. Erdogan, W. Friedt and F. Ordon, 1995: Potential and limitations of isozymes for chromosomal localization of resistance genes against barley mild mosaic virus (BaMMV). Euphytica 82: 25-30.
Ordon, F., E. Bauer, W. Friedt and A. Graner, 1995: Marker-based selection for the ym4 BaMMV-resistance gene in barley using RAPDs. Agronomie 15, 481-485.
Sobral, B.W.S. and R.J. Honeycutt, 1993: High output genetic mapping of polyploids using PCR-generated markers. Theor. Appl. Genet. 86, 105-112.
Weyen, J., E. Bauer, A. Graner, W. Friedt and F. Ordon, 1996: RAPD-mapping of the distal portion of chromosome 3 of barley, including the BaMMV/BaYMV resistance gene ym4. Plant Breeding 115, 285-287.