The discontinuous system of polacrylamide gel (PAAG) electrophorosis was used to separate the esterase isozymes (Netsvetaev, 1992). The tray buffer, pH 8.3, included Tris, EDTA, and boric acid. PAAG electrophoresis for ß-amylase was accomplished with the tris-glycine system, pH 8.3 (Netsvetaev, 1993). Esterase extract from 7 to 8 day-old seedlings was obtained in a 0.024 M sodium bicarbonate. Mature dry seeds were used for extraction of ß-amylase. Flour from the endosperm was poured into a solution consisting of 0.024 M sodium bicarbonate, 0.01 M dithiothreitol, 0.6 M sucrose, and bromophenol blue for coloring. After centrifugation, the supernatant was used for electrophoretic separation of ß-amylase isozymes.
DNA was obtained from 7 to 8 day-old seedlings. The leaf blade from one seedling was crushed with a glass rod in an Eppendorf tube that contained 0.5 ml of lysing solution (0.1 M Na3EDTA, 10 mM tris-HCl pH 8.5, 100 mM NaCl, 1% SDS, 0.1% Triton X-100, and 100 µg/ml proteinase K). For protein destruction, the samples were treated with this solution for 2 hours at 55°C. Peptides were dissolved in volumes of a chloroform-ispropenthanol mixture. The DNA was precipitated with a 0.5 volume of isopropanol. The PCR was conducted according with the method described by Sivolap and Kalendar' (1994). The reaction volume was 20 µl and contained 16.6 mM (NH/4/)/2/SO/4/, 4 mM MgCl/2/, 0.2 mM each dNTP, 0.01% w/w Tween-20, 57 mM Tris-HCl (pH 8.8), 200 nM primer P6, 20 ng DNA, and 1 unit Taq polymerase. Samples were overlaid with light mineral oil Bajol F (Serva). The first cycle of PCR was carried out under the following conditions: 2 min. at 94°C, 2 min. at 42°C, and 2 min. at 72°C. Conditions for cycles 2 to 4 were 1 min. at 94°C, 2 min. at 42°C, and 2 min. at 72°C. Cycles 5 to 37 had the parameters: 1 min. at 94°C, 1.5 min. at 52°C, and 2 min. at 72°C. This was followed by a final incubation at 72°C for 7 min. The PCR products were electrophoresed in 2% agarose gel.
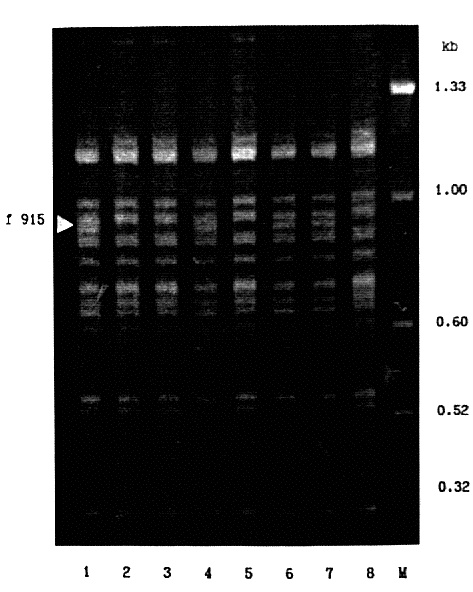
The genotypes of the parents were: Odessky 115 - Est1Ca, Est5Pi, Bmy1Ar, and absence of the PCR 915B fragment (f915); Golf - Est1Pr, Est5Te, Bmy1Br, and presence of f915. Data for linkage estimates between genetic and molecular markers in F->inf. Odessky 115 X Golf lines are summarized in Table 1. Since no recombinants were found, the f915 locus showed complete linkage with the ß-amylase locus, Bmy1. The other genetic factors were inherited independently of f915 (Table). Thus, the f915 locus is located at (or near) the Bmy1 locus in the long arm of chromosome 4. Presence of the f915 fragment was associated with the slow ß-amylase 1 isozyme (Fig. 2). which is determined by the Bmy1Br allele.
References
Netsvetaev, V.P. 1992. Identification of barley leaf esterases 11, 12 and their control. Genetika 28:105-119 [In Russian].
Netsvetaev, V.P. 1993. Location of the ß-amylase locus (Bmy1) in chromosome 4 of barley. Cytology and Genetics, Kiev 27:74-78. [In Russian].
Netsvetaev, V.P., and A.A. Sozinov. 1984. Location of a hordein G locus, Hrd G,on chromosome 5 of barley. BGN 14:4-6.
Sivolap, Yu.M., and R.N. Kalendar'. 1994. PCR with arbitrary primers for detection of genetic polymorphism in barley. Genetika (In press) [In Russian].
Table 1. Linkage data between PCR fragment (f915) and the Bmy1, Est1, and Est5 loci in a cross of barley cultivars (F->inf. Odessky 115 X Golf).
_______________________________________________________________
Genetic factors No. of F->inf. families
_______________ _____________________
Pheno- Total Recombination
A X B types BB bb n X²L %
_______________________________________________________________
Bmy1 X f915 AA 65 0
aa 0 75 140 140.0 0.0 ± 0.3
Est1 X Est5 AA 32 23
aa 16 22 93 2.4 Independent
Est1 X f915 AA 25 34
aa 22 16 97 2.3 Independent
Est5 X f915 AA 21 30
aa 26 20 97 2.3 Independent
_______________________________________________________________

