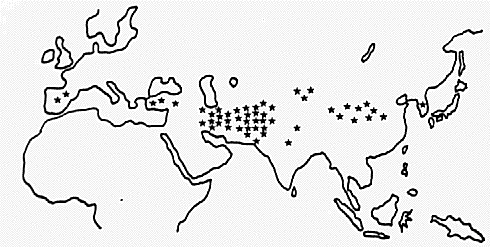
Fig. 1. Geographical distribution of the varieties with negative reaction to phenol
K. Takeda and C. L. Zhang
Research Institute for Bioresources, Okayama University, Kurashiki, Japan
According to the preliminary experiment, the grains or spikes of barley were heated with 1 % phenol solution, adjusted to pH 6, for 2 or 3 days at room temperature to check the reaction. The reaction of awn was the sharpest in all parts of the plant.
Positive reaction to phenol may be the prototype of the trait, because all the wild relatives examined showed a positive reaction.
In most of the cultivated varieties, awns or grains were clearly stained by the phenol solution, but in some of the varieties the reaction was not clear. Those varieties were examined repeatedly and finally 51 varieties showing negative reaction to phenol were detected. These varieties mainly distributed along the so-called "Silk road". One variety with negative reaction was found in the Korean peninsula and two varieties existed in the Iberian peninsula, but none was in the Europe, except Spain, New Continent and Africa (Fig. 1). This limited distribution of the varieties with negative reaction to phenol indicates that it is a rather new mutant which appeared in the Middle East and was expanded in distribution by mankind.

Fig. 1. Geographical distribution of the varieties with negative reaction to
phenol
A total of 18 crosses were made between 8 linkage testers with positive phenol reaction and 3 varieties with negative reaction.
All of the F/1/ plants showed positive reaction, suggesting the gene is dominant. The F/2/ plants were raised to check the marker characters and spikes of the plants were harvested test the phenol reaction. A total of 3,704 F/2/ Plants from 18 crosses segregated into the 3 to 1 ratio (Table 1). Thus, the phenol reaction in barley is controlled by a single dominant gene. In case of rice plant, the gene for phenol reaction is named Ph, however in barley we propose a gene symbol Phr for preventing possible confusion with Ph (paring homoeologus) gene in wheat where a similar locus may be found in the genus Hordeum.
Table 1. Segregation of phenol reaction in the F/2/ generation (total of 18 pop.)
___________________________________________________________
Positive Negative Total X²(3:1) p
___________________________________________________________
Obs. 2,779 925 3,704 0.001 >0.9
Exp. 2,778 926
___________________________________________________________
Analyzing the joint segregation of Phr and marker genes, Phr was
independent of marker genes belonging to chromosomes 1 and 3 to 7 : namely,
n (naked karyopsis) and br (brachytic plant) on chromosome 1,
als (absent lower laterals), uz (uzu or semi-brachytic growth)
and al (albino lemma) on chromosome 3, K (hooded lemma), gl-3
(glossy seedling-3) and Bl (blue aleurone) on chromosome 4, trd
(third outer glume) and B (black lemma and pericarp) on chromosome
5, o (orange lemma) on chromosome 6 and s (short haired rachilla)
on chromosome 7.As shown in Table 2, the Phr gene was linked with three marker genes located on the chromosome 2: namely, e (wide outer glume), V (two-rowed) and li (ligule-less). The arrangement of the four genes on the chromosome 2 is shown in Fig. 2.
Table 2. Linkage analysis among the Phr and three marker genes on the chromosome 2 in OUI677 x OUL017 F/2/ population
__________________________________________________________ Genes (A:B) A-B- A-bb aaB- aabb Total X²L RCV(%) __________________________________________________________ Phr : e 205 78 94 17 394 7.04** 39.5 ±4.2 Phr : li 197 86 106 5 394 31.46** 21.8 ±4.8 Phr : V 242 41 48 63 394 82.84** 24.7 ±2.6 li : V 209 94 81 10 394 14.40** 32.3 ±4.4 li : e 235 68 64 27 394 1.90 44.7 ±3.5 e : V 200 99 90 5 394 29.24** 22.2 ±4.7 __________________________________________________________**: Significant at the 1% level.
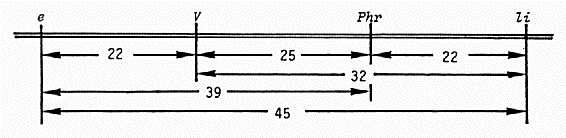
Fig. 2. Linkage relation of the Phr and other marker genes on the
chromosome 2.
The Phr gene is a new marker gene on the long arm of chromosome 2. Phenol reaction is an additional key character for investigating the phylogeny of the barley. Physiological studies of the trait are under way.
Reference
Takasugi, S. 1937. Varietal variation in coloring of seeds with the phenol, sulfuric acid and caustic potash solutions in Japanese barley. Agriculture and Horticulture 12: 1101-1105.(In Japanese)