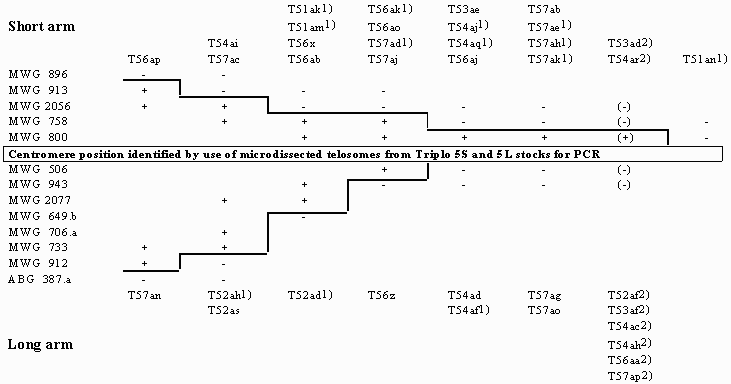
+/-: RFLP probe-specific PCR fragment present/absent. Empty fields: not tested
Using the polymerase chain reaction (PCR) with sequence-specific primers and template DNA from microisolated translocation chromosomes of Vicia faba, Macas et al. (1993) presented a new strategy to physical gene localization in plants. Adaptation of this technique to barley (Sorokin et al. 1994) allowed us to construct for the first time a cytogenetically based physical RFLP map of a barley chromosome. Based on four suitable translocations with breakpoint positions previously localized to relatively short segments of Giemsa-banded somatic metaphase chromosomes (Marthe and Künzel 1994) and an appropriate selection of RFLP probes, the 37 RFLP loci mapped within the Igri/Franka derived linkage group 5 (Graner et al. 1994) could be assigned to five defined subregions of chromosome 5 (Künzel et al. 1995).
Extending the same PCR technique to additional translocation chromosomes, a total of 37 breakpoints have been exactly positioned between the RFLP loci of linkage group 5 (Table 1). With two exceptions, T51an and T57ad, the PCR results fit to the physical data obtained by karyotyping (Table 2). Since the physical position of RFLP loci was confirmed by repeated PCR experiments, the karyotypically determined positions for T1-5am and T5-7ad are erroneous. Based on the PCR results these breakpoint positions were revised.
Figure 1 illustrates the location of the 37 breakpoints in relation to the RFLP and the physical map. Most of the breakpoints (31) are clustered within a region of about 15 cM around the centromere. This relatively small part of the genetic map (about 10%) corresponds to about 60% of the total chromosome lenght and represents an area of suppressed meiotic recombination. Exceptionally, one of the few breakpoints located outside this region (T57an) was positioned between the outermost two loci of the long arm.
The localization of a large number of breakpoints exactly between the linear array of genetic markers allowed to refine the breakpoint positions determined previously by karyotype analysis for several translocations. This is especially the case if breakspoints, karyotypically assigned to overlapping segments, occur between the same two flanking markers as specified in Table 2 for 20 of the investigated 37 translocations involving chromosome 5.
Acknowledgement
This research was supported by the Federal Ministry for Research and Technology (Grant No. 0319960B).
Tab. 1. Integration of 37 translocation breakpoints of chromosome 5 into the Igri/Franka derived RFLP linkage map. PCR results with RFLP probe-specific primers and DNA of microisolated translocation chromosomes

+/-: RFLP probe-specific PCR fragment present/absent. Empty fields: not
tested
¹) Only one of the two reciprocally interchanged chromosomes distinct enough for microisolation to be used for PCR
²) Insertion based on breakpoint positions determined by karyotyping (Marthe and Künzel 1994)
Tab. 2. Cytological localization of breakpoints in 20 reciprocal translocations as improved by ordering the breakpoints of chromosome 5 into the linear range of RFLP markers of linkage group 5
Breakpoints assigned to segments
defined in mGN¹
Determined by Refined by
T-Line Chromosome karyotyping PCR results
__________________________________________________________
T1-5am T15am 1S:27 to 36 29 to 36
T51am 5S:23 to 32 25 to 32
T1-5an* T15an 1S:(27 to 36) 3 to 14
T51an 5S:(24 to 33) 0 to 11
T3-5ad T35ad 3L:20 to 27 21 to 27
T53ad 5S:10 to 17 11 to 17
T3-5ae T35ae 3L:55 to 82 56 to 77
T53ae 5S:10 to 37 11 to 32
T4-5af T45af 4L:36 to 69 36 to 66
T54af 5L:29 to 62 29 to 59
T4-5ai T45ai 4L:28 to 40 28 to 36
T54ai 5S:38 to 50 38 to 46
T4-5aj T45aj 4L:28 to 53 28 to 35
T54aj 5S:25 to 50 25 to 32
T4-5aq T45aq 4S:13 to 14 14
T54aq 5S:10 to 11 11
T5-6x T56x 5S:10 to 32 25 to 32
T65x 6S:19 to 41 34 to 41
T5-6ab T56ab 5S:10 to 50 25 to 46
T65ab 6L:22 to 62 37 to 58
T5-6aj T56aj 5S:10 to 12 11 to 12
T65aj 6S: 8 to 10 9 to 10
T5-6ak T56ak 5S:10 to 50 25 to 46
T65ak 6L:26 to 66 41 to 62
T5-6ao T56ao 5S:10 to 48 25 to 46
T65ao 6L:32 to 70 47 to 68
T5-6ap T56ap 5S:10 to 46 38 to 46
T65ap 6L:34 to 70 62 to 70
T5-7ac T57ac 5S:24 to 50 30 to 46
T75ac 7L:22 to 48 28 to 44
T5-7ad* T57ad 5S:(11 to 17) 25 to 46
T75ad 7L:(10 to 16) 24 to 45
T5-7ah T57ah 5S:10 to 35 11 to 32
T75ah 7L:35 to 60 36 to 57
T5-7aj T57aj 5S:19 to 40 25 to 40
T75aj 7S:22 to 43 28 to 43
T5-7ak T57ak 5S:12 to 50 12 to 32
T75ak 7L:22 to 60 22 to 42
T5-7ao T57ao 5L:29 to 62 29 to 59
T T75ao 7L:25 to 58 25 to 55
_________________________________________________________
¹ According to Jensen and Linde-Laursen (1992)
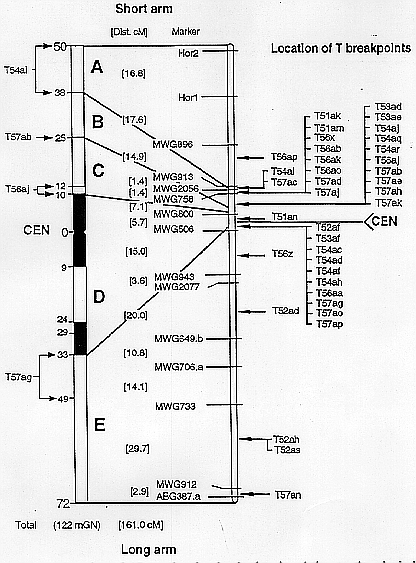
Fig. 1. Location of 37 translocation breakpoints in relation to the
physical and the Igri/Franka derived RFLP map for chromosome 5. Left-hand:
Physical map (idiogram) with measurements in milliGeNomes (mGN) according to
Jensen and Linde-Laursen (1992). Breakpoints used for map construction are
marked by arrows; single arrow for the precisely located breakpoint of T57ab,
cramped arrows indicate the segments within which the breakpoints of T54ai,
T56aj and T57ag are located. Right-hand: Skeletal genetic map indicating the
location of breakpoints within the linear array of RFLP markers. Corresponding
regions of the genetic and physical map are indicated by lines.
References
Graner, A., E. Bauer, A. Kellermann, S. Kirchner, J.K. Muraya, A. Jahoor, and G. Wenzel. 1994. Progress of RFLP-map construction in winter barley. BGN 23: 53-59.
Jensen, J., and I. Linde-Laursen. 1992. Statistical evaluation of length measurements on barley chromosomes with a proposal for a new nomenclature for symbols and positions of cytological markers. Hereditas 117: 51-59.
Künzel, G., A. Sorokin, and F. Marthe. 1995. Progress in relating genetic to physical distances of marker sequences on microisolated barley chromosomes. In: "Kew Chromosome Conference IV", Proc. 4th Kew Chromosome Conference, submitted.
Macas, J., W. Weschke, H. Bäumlein, U. Pich, A. Houben, U. Wobus, and I. Schubert. 1993. Localization of vicilin genes via polymerase chain reaction on microisolated field bean chromosomes. Plant J. 3: 883-886.
Marthe, F., and G. Künzel. 1994. Localization of translocation breakpoints in somatic metaphase chromosomes of barley. Theor. Appl. Genet. 89: 240-248.
Sorokin, A., F. Marthe, A. Houben, U. Pich, A. Graner, and G. Künzel. 1994. Polymerase chain reaction mediated localization of RFLP clones to microisolated translocation chromosomes of barley. Genome 37: 500-555.