

Introduction
In the past, the localisation of loci for quantitative traits (QTLs) seemed inaccessible. One main reason was the lack of dense linkage maps with markers for individual chromosomes of a plant species having nopleitropic effect on the trait of interest. Looking at practical breeding, on the one hand, a lot of the traits of agronomic importance are of quantitative characer but on the other hand the selection, for these traits is complicated by the number of genes involved in it and/or the environmental effects. Hence, the discovery of molecular markers with all their advantages, as their almost unlimited availability, the clear distinction between homozygous and heterozygous individuals and the fact, that in most cases, they are sequences without any pleiotropic influence to any trait, offered the opportunity to study the inheritance of quantitative traits. With the development of dense genetic maps in several plant species of agronomic interest (Helentjaris, 1987, Liu and Tsunewaki 1991, Gebhardt et al. 1991), the initial step was done to map QTLs. Consequently, lately localisations of quantitative traits for many crop plants such as tomato (Kinzer et al. 1990), maize (Melchinger et al. 1992) and barley (Hayes et al. 1993) were published. Furthermore, molecular markers would make it possible to find the 'optimal' allele combination for a quantitative trait by marker assisted selection (MAS) in a very early stage of the breeding process (Tanksley and Rick, 1980, Zhang and Smith 1992).
Material and Methods
A total of 250 doubled haploid lines were produced from a cross of the two row winter barley varieties 'Igri' and 'Danilo'. The DH-lines originating from anther culture were selfed eight times before including in the present investigation. These lines were raised in field experiments on two locations during three years. From these experiments, the following observations were evaluated: infection severity of Rhynchosporium Cleaf spots'), infection severity of powdery mildew, lodging, stalk breaking, ear breaking and field condition before harvesting. All data where scored from 1 to 9. where '1' is always the most 'positive' and 9 the most 'negative'.
The plant height and the heading date, as days after the first of May, were also obtained, Moreover, kernel weight, kernel length, kernel thickness, kernel shape (quotient of kernel length by kernel thickness) and kernel yield were examined after harvesting.
The plant genomic DNA was isolated according to the CTAB procedure (SaghaiMaroof et al., 1984, modified), digested with the restriction enzymes BamHI, EcoRI, EcoRV, HindIII, SacI and XbaI, separated electrophoretically and transferred to nylon membranes (Southern, 1975). The probes were developed in a co-operation project of the Department of Agronomy and Plant Breeding in Weihenstephan, the institute for Resistance Genetics in Granbach and the Institute of Botany of the Ludwig-Maximilians-University in Munich and were labelled with 32P by the Random Primed Labelling Method (Feinberg and Vogelstein, 1983).
The RFLP-based map was established using the MAPMAKER 3.0 software (Lander et al., 1987). The detection of linkage between the probes and the QTLs was performed using analysis of variance (SPSS for Windows procedure MANOVA, SPSS Corp.) and MAPMAKER QTL 1.1 (Paterson et al. 1988).
Results and Discussion
In this cross, only a low degree of polymorphism polymorphism's could be observed: 50 out of 431 actually examined probes showed band differences i.e. 11,6%. With these probes, 54 loci could be mapped UH: one locus, 2H: 7 loci, 3H: 11 loci, 4H: 7 loci, 5H: 10 loci, 6H: 10 loci, 7H: 8 loci). The different linkage groups were associated to the chromosomes by using information of previously published RFLP maps (Graner et al., 1991).
The first examination of data was accomplished by an analysis of variance (ANOVA) for every probe and every trait, including the variance explained by the probe, by the environment and by the probe × environment. Only some of the probes showed weakly significant probe × environment interaction.
The main examination of the data was performed by MAPMAKER QTL. There were no striking differences between the two types of data exploration. Many of the probes with very high significant influence on the QTLs ((α < 0,001) in the ANOVA analysis did not reach a LOD of even 2 in the multi-point maximum-likelihood analysis by MAPMAKER QTL, Therefore, it was decided to accept QTLs with an LOD higher than 2 in the present investigation.
The results of the analysis are presented in table 1. The part of total variance explained by the genetic variance (Vg/Vt) and the part of the complete variance, corrected for systematic errors (environment and probe × environment) explained by the genetic variance (Vg/Vc) are revealing a kind of 'aptitude' of this trait for the QTL analysis. A large non systematic error would turn it difficult or even impossible to detect quantitative loci. Very high quotients can be observed for 'heading date' and the kernel parameters. The very low quotient for 'Powdery Mildew' is explained by the poor epidemiology in these three years.
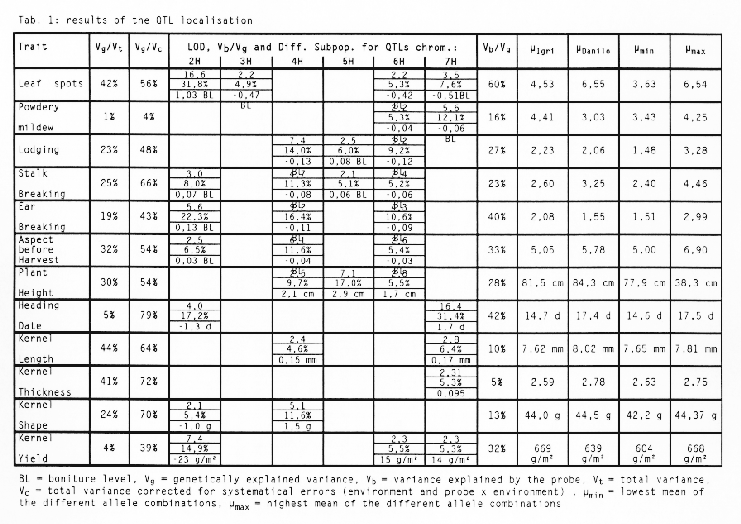
For every QTL the table 1 displays the location of the locus on the chromosomes, the LOD, the explained part of the genetical variance and the differences in the trait between the two subpopulations constituted by the alleles of the probe. A positive sign of the difference denotes, that the population with the allele from the 'Igri' parent shows a lower value than the population with the 'Danilo' allele and vice versa for the negative sign of the difference. QTLs which explain more than 30% of the genetic variance can be found on chromosome 2H for 'leaf spots' and on chromosome 7H for 'heading date'. The QTL for kernel yield detected on chromosome 2H can be explained completely by the QTL for 'leaf spots' on the same chromosome, as can be shown by an ANOVA with 'leaf spots' as a covariate. This fact demonstrates, that localising traits with high complexity and interdependencies with numerous other traits should be accomplished very carefully.
The explained part of the genetic variance (Vq/Vg) and the highest (µmax) and lowest means (µmin) of the different combination of alleles were calculated taking into account all QTLs localised for a particular trait. There were no significant pleiotropic effects between the QTLs for one trait. The combination of alleles with highest scores always showed the highest scores for the trait and vice versa. For almost every trait these combinations have not been identified in the parents. Transgression effects of variable extent are occurring in the allele-combination groups described above compared to the mean of the parents (µIgri and µDanilo).
Acknowledgements
The authors thank Dr. B. Foroughi-Wehr for providing the DH mapping population and E. Dietzman for delivering the field data. The helpful technical assistance of A. Wegmann ist greatly appreciated. This study was supported by the Federal Ministry for Research and Technology (Grant No. 0318990G)
References
Feinberg, A.P. und B. Vogelstein (1983): A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132: 6-13
Gebhardt C., Ritter E., Schachtschabel U., Kaufmann H., Tanksley S.D. und F. Salamini Barone A. , Debener T., Walkemeier B., Thompson R.D., Bonierbale M.W., Ganal M.W., (1991) RFLP maps of potato and their alignment with the homoeologous tomato genome. Theor. Appl. Genet. 83:49-57
Graner, A., Jahoor, A., Schondelmaier, J., Siedler, H., Pillen, K., Fi schbeck, G., Wenzel , G. und R. G. Herrmann ( 1991 ): Construction of an RFLP map of barley. Theor. appl. Genet. 83: 250-256.
Hayes, P.M, Liu, B.H., Knapp, S.J., Chen, F., Jones , B., Blake, Franckowiak, J., Rasmusson, D. , Sorells, M., Ullrich, S.E., Wesenberg, D. und A. Kleinhofs (1993): Quantitative trait locus effects and environmental interaction in a sample of North American barley germplasm. Theor. Appl. Genet. 87: 392-401.
Helentjaris T (1987) A genetic linkage map for maize based on RFLPs. Trends Genet. 3: 217-221
Kinzer, S.M., Schwager, S.J. und M.A. Mutschler (1990): Mapping of ripening-related or -specific cDNA clones of tomato (Lycopersicon esculentum). Theor. Appl. Genet. 79: 489-496.
Lander E., Green, P., Abrahamson, J., Barlow, A., Daley, M., Lincoln, S. und L. Newburg (1987): MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174-181.
Liu Y.G, and Tsunewaki K. (1991) Restriction fragment length polymorphism (RFLP) analysis in wheat. II. Linkage maps of the RFLP sites in common wheat. Jpn. J. Genet. 66 : 617-633
Melchinger, A.E., Bopppenmaier, J., Dhillon, B.S., Pollmer, W.G. und R.G. Herrmann (1992): Genetic diversity for RFLPs in European maize inbreds: II. Relationsships to performance of hybrids within versus between heterotic groups for forage traits. Theor. Appl. Genet. 84: 672-681.
Paterson, A.H., Lander, E.S., Hewitt, J.D., Peterson, S., Lincoln, S.E. und S.D. Tanksley (1988): Resolution of quantitative traits into Mendelian factors by using a complete linkage map of restriction fragment polymorphism. Nature 225: 721-726
Saghai-Maroof M.A., Soliman K.M., Jorgensen, R.A. und RW. Allard (1984): Ribosomal spacerlength polymorphisms in barley: Mendelian inheritance, chromosomal location and population dynamics. Proc. Natl. Acad. Sci. USA 81: 8041-8018
Southern, E.M. (1975): Detection of specific sequences among DNA fragments seperated by Gel Electrophoresis. J. Mol. Biol. 98: 503-517.
Tanksley S.D. and Rick, C.M. (1980): Isozymic Gene Linkage Map of Tomato. Applications in Genetics and Breeding. Theor. Appl. Genet. 57: 161-170.
Zhang W. und Smi th , C . ( 1992) : Computer simulation of marker-assisted selection utilising linkage desequilibrium. Theor. Appl. Genet. 83: 813-820.