

The first duplication of chromosome segments in barley was described by Hagberg (1962). Later the following papers have been published: Hagberg et al. (1987, 1991); Hagberg et al. (1983); Hagberg et al. (1974, 1978). All of these duplication lines, except one (D49) which was lost, are kept in a "working collection" at Svalöv and used for cytogenetic research. In 1992 the lines will be multiplied and also included in the Nordic Gene Bank (NGB), Alnarp, Sweden. Seed samples are available upon request. Requests should preferably be sent to the coordinator.
The duplications in the collection were obtained from the segregating progenies of crosses between translocation lines (= chromosome segment interchanges) involving the same two pairs of chromosomes. To form viable duplications, the relative position of the two break points in the respective translocation chromosome must fulfill specific requirements, which are described in the papers quoted above and by Burnham and Hagberg (1956). To find the combination of translocations which can yield duplications, you need a supply of translocations which are well identified. Table 1 shows the total number of chromosome breakpoints in the translocation collection at Svalöv and their distribution among the chromosomes involved. This collection of translocations has been included earlier in the NGB stocks. For further information on the international collection of translocations is referred to the coordinators of translocation stocks, Drs. G. Künzel and R. T. Ramage.
Thus, the basic material in the production of duplications is the translocations mainly produced by ionizing radiation. Of the 692 translocations included in Table 1, those involving chromosomes carrying a satellite (chromosome 6 and 7) are the most suitable for cytological studies. The results that have been published comprise T6-7, T5-6, and T5-7 translocations.
The 29 T5-6 translocations were crossed diallelically to obtain information about their relative break positions in respective chromosome and to produce duplications. The duplications D5-6 found were named D201-D203.
The 32 T5-7 translocations were diallelically crossed and the D5-7 lines found were named D101-D103. The vitality and fertility of most duplications involving chromosome 5 was low. This is the reason there are only a few lines available.
Among T6-7 translocations, 51 were used for diallel crosses. The crosses yielded 51 duplication lines which are listed in Table 2. They were assigned numbers in the same order as they were identified: from Dl to D52 (one number was lost, D49).
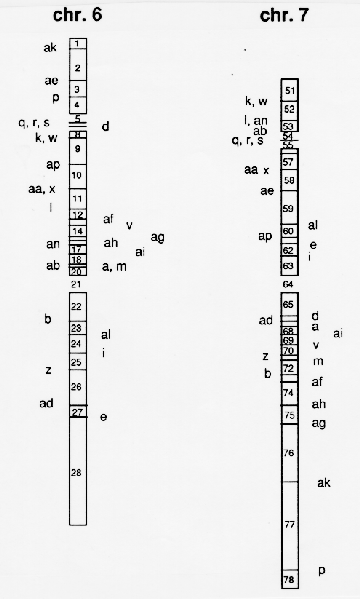
The first D6-7 was obtained in the F2 generation of the cross T6-7a x T6-7d. It is found in Table 2 as D1 followed by 1) a bracket with the letters of the T-lines used in the actual cross and 2) numbers referring to the chromosome segments involved in the duplication. The segment numbers are explained in Fig. 1: chromosome 6 and 7 have been divided into segments by indicating the various translocation breakpoint positions determined by cytogenetic observations of the translocation lines. Chromosome 6 has thus been divided into segments 1 to 28. The corresponding breakpoints in chromosome 7 are giving the segments 51 to 78.
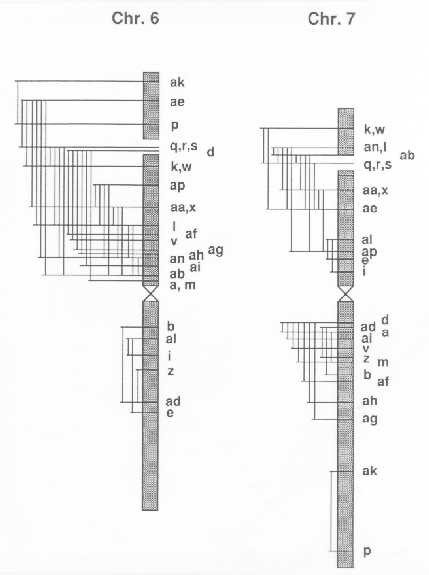
The breakpoints tentatively located with the method described are indicated in Fig. 1 by the letters ak, ae, p, etc., which are abbreviations for T6-7ak, T6-7ae and T6-7p etc. Also the number of the actual segments are shown. In Fig. 2 the positions of the duplicated segments in 51 D6-7 lines available are indicated for direct comparison of the segments involved.
| Chromosome | 7 | 6 | 5 | 4 | 3 | 2 | 1 | Sum of T-breaks in each chromosome |
|---|---|---|---|---|---|---|---|---|
| 1 | 31 | 37 | 36 | 30 | 40 | 28 | - | chr. 1:202 |
| 2 | 33 | 38 | 31 | 32 | 33 | - | chr. 2:195 | |
| 3 | 33 | 27 | 28 | 35 | - | chr. 3:196 | ||
| 4 | 29 | 28 | 35 | - | chr. 4:185 | |||
| 5 | 32 | 29 | - | chr. 5:187 | ||||
| 6 | 51 | - | chr. 6:210 | |||||
| 7 | - | chr. 7:209 |
| Duplication | Origin | Segment duplicated in | |
|---|---|---|---|
| chr. 6 | chr. 7 | ||
| D41 | (ap x aa) | 10 | 58-60 |
| D15 | (ap x x) | 10 | 58-60 |
| D12 | (x x 1) | 11 | 53-57 |
| D11 | (aa x 1) | 11 | 53-57 |
| D9 | (ap x 1) | 10-11 | 53-60 |
| D8 | (1 x an) | 12-16 | |
| D50 | (ap x an) | 10-16 | 53-60 |
| D14 | (x x an) | 11-16 | 53-57 |
| D18 | (aa x an) | 11-16 | 53-57 |
| D16 | (x x ab) | 11-18 | 54-57 |
| D19 | (aa x ab) | 11-18 | 54-57 |
| D10 | (1 x ab) | 12-18 | 53 |
| D23 | (ab x ap) | 10-18 | 54-60 |
| D13 | (x x ae) | 3-10 | 58 |
| D17 | (aa x ae) | 3-10 | 58 |
| D6 | (ab x ae) | 3-18 | 54-58 |
| D5 | (an x ae) | 3-16 | 53-58 |
| D7 | (1 x ae) | 3-11 | 53-58 |
| D22 | (q x ab) | 6-18 | 54 |
| D21 | (r x ab) | 6-18 | 54 |
| D20 | (s x ab) | 6-18 | 54 |
| D52 | (q x an) | 6-16 | 53-54 |
| D26 | (r x an) | 6-16 | 53-54 |
| D51 | (s x an) | 6-16 | 53-54 |
| D48 | (q x 1) | 6-9 | 53-54 |
| D44 | (r x 1 | 6-9 | 53-54 |
| D43 | (s x 1) | 6-9 | 53-54 |
| D24 | (q x ae) | 3-5 | 55-58 |
| D25 | (r x ae) | 3-5 | 55-58 |
| D29 | (s x ae) | 3-5 | 55-58 |
| D45 | (q x k) | 6-8 | 52-54 |
| D28 | (r x k) | 6-8 | 52-54 |
| D30 | (s x k) | 6-8 | 52-54 |
| D46 | (q x w) | 6-8 | 52-54 |
| D27 | (r x w) | 6-8 | 52-54 |
| D47 | (s x w) | 6-8 | 52-54 |
| D3 | (k x ae) | 3-8 | 52-58 |
| D2 | (w x ae) | 3-8 | 52-58 |
| D4 | (p x ak) | 2-3 | 77 |
| D1 | (a x d) | 7-19 | 66-67 |
| D35 | (d x m) | 7-9 | 68-71 |
| D40 | (a x m) | 66-71 | |
| D31 | (d x v) | 7-13 | 68-89 |
| D32 | (d x ai) | 7-17 | 68 |
| D34 | (d x af) | 7-12 | 68-73 |
| D36 | (d x ah) | 7-15 | 68-74 |
| D33 | (d x ag) | 7-14 | 68-75 |
| D42 | (e x al) | 24-27 | 60-61 |
| D37 | (i x al) | 24 | 60-62 |
| D38 | (b x ad) | 23-26 | 67-72 |
| D39 | (z x ad) | 26 | 67-70 |
References:
Burnham, C. R., and A. Hagberg. 1956. Cytogenetic notes on chromosomal interchanges in barley. Hereditas 42:467-482.
Hagberg, A. 1962. Production duplications in barley. Hereditas 48:243-246.
Hagberg, A., and G. Hagberg. 1987. Some vigorous and productive duplications in Barley. In: Barley Genetics V. Proceedings of the 5th Int. Barley Genet. Symp. Okayama. pp.423-426.
Hagberg, A., and P. Hagberg. 1991. Production and analysis of chromosome duplications inbarley. In: Chromosome Engineering in Plants, Part A. Ed. Gupta and T. Tsuchiya. Elsevier pp: 401-410.
Hagberg, G., A. Hagberg, and T. Sodkiewicz. 1983. Duplication of chromosome segments in barley chromosomes 6 and 7. BGN 13:32-35.
Hagberg, P. 1974. Duplication of tandem satellites. Progress report. BGN 4:29-31.
Hagberg, P., and A. Hagberg. 1978. Segmental interchanges in barley III. Translocations involving chromosomes 6 and 7 used in production of duplications. Z. Pflanzenzüchtung 81:111-117.