

In eukaryotes the ribosomal genes (rRNA; 18S-5.8S-26S) are organized as families of tandemly repeated units located at one or a few chromosomal sites (Flavell, 1986). In barley (Hordeum vulgare), chromosomes 6 (6H) and 7 (SH) carry the major sites of rRNA genes. Here we have used a sensitive fluorescent in situ hybridization technique to map the physical location and relative number of copies of the rRNA genes on the chromosomes of Hordeum Vulgare cv. Sultan.
In situ hybridization based on the method described by Heslop-Harrison et al. (1991) and Leitch et al. (1991) was carried out on chromosomes prepared by dropping fixed root tip cells onto clean microscope slides. pTa71, a 9 kb EcoRl fragment of rDNA isolated from Triticurn aestivum (Gerlach and Bedbrook, 1979), recloned into pUC19, and provided by R. B. Flavell and M. O'Dell, JI Centre, Norwich, was used as the probe. It contains the coding sequences for the 18S, 5.8S, and 26S genes and non-transcribed spaced sequences. pTa71 was labelled with digoxigenin-11-dUTP (Boehringer Mannheim) by nick translation.
In total, thirteen complete metaphase cells were examined in detail following in situ hybridization. Signal was recorded where sites were present on both chromatids. All thirteen spreads showed the four major rDNA sites and additional minor sites. Six cells showed ten sites, four showed nine sites and one, seven sites. In cells where fewer than ten sites were present, additional hybridization sites were visible on single chromatids. Two further cells had eleven sites, but both the shape and relative strength of one site indicated that it was an artifact. Chromosomes were identified by C-banding following in situ hybridization by comparison with standard C-banded karyotypes of barley (Linde-Laursen, 1978).
The additional sites of the rDNA sequences detected by in situ hybridization were located on the short arms of chromosomes 5 (1H), 1 (7H), and 2 (2H). The presence and position of the rDNA sites on the short arm of chromosomes 1 (7H) and 2 (2H) were confirrned by repeating the in situ hybridization experiment on barley lines telotrisomic for the short arm of chromosome 1 (7H) and 2 (2H) (Singh and Tsuchiya, 1977). Measurements indicated that the average positions of the signal for chromosomes 5 (1H), 1 (7H), and 2 (2H) were 37, 38, and 52%, respectively, along the labelled arm from the centromere. An idiogram showing both the position of the in situ hybridization signal and the C-bands for one of the six cells showing ten in situ hybridization sites is given in Fig. 1.
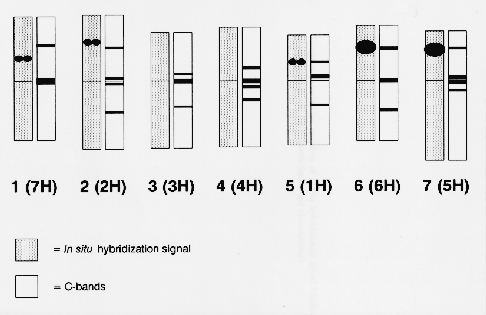
The in situ hybridization signal on chromosome 5 (1H) was stronger than that on the other two chromosomes carrying signal. Although in situ hybridization is not a fully quantitative technique, the strength of hybridization signal is approximately related to the copy number of genes when they have similar structures (Appels et al., 1980; Maluszynska and Heslop-Harrison, 1991). It is estimated that the site on chromosome 5 (1H) corresponds to between 50 and 100 copies of the rRNA sequence while the weaker signals detected on chromosomes 1 (7H) and 2 (2H) correspond to between five and ten copies. These additional rDNA sites have not previously been detected by Southern hybridization analysis, probably because the hybridization signals from the two major sites [6 (6H) and 7 (5H)] masked weaker hybridization signals produced by the additional sites, as suggested by Mukai et al. (1991). Similarly, the extra sites could not be mapped using DNA extracted from barley chromosome addition lines to wheat because of excess numbers of wheat rRNA genes, and the possible presence of a few copies of variants within the major sites.
rRNA genes are known by the gene symbol Nor without regard to their activity, capacity to organize a nucleolus (Mukai et al., 1991). Based on the nomenclature of previous rDNA sites, the number assigned is related to the temporal order of discovery on each chromosome group (homoeologous chromosome group 1 = Norl; group 6 = Nor2; group 5 = Nor3; and group 7 = Nor4). The two major rDNA sites in barley are called Nor-H2 on chromosome 6 (6H) and Nor-H3 on chromosome 7 (SH), or alternatively Rrnl and Rrn2. We designate the newly identified sites on barley chromosomes 5 (1H) and 1(7H), Nor-HI and Nor-H4, respectively. The site on chromosome 2 (2H) represents the first designation of rDNA sequences on homoeologous group 2 chromosomes, so we call this site Nor-H5, and propose that further rDNA sites identified on homoeologous group 2 chromosomes are numbered Nor5.
Acknowledgements:
We thank Dr. Trude Schwarzacher for helpful discussions, assistance with the chromosome dropping, and C-banding methods. And we are grateful to Dr. Ib Linde-Laursen for assisting with the identification of the C-banded chromosomes. We thank AFRC PMB grant 569/112 for support.
References:
Appels, R., W. L. Gerlach, E. S. Dennis, H. Swift, and W. J. Peacock. 1980. Molecular and chromosomal organization of DNA sequences coding for the ribosomal RNAs in cereals.Chromosoma 78:293-311.
Flavell, R. B. 1986. The structure and control of expression of ribosomal RNA genes. Oxford Surveys of Plant Molecular and Cell Biology 3:252-274.
Gerlach, W. L., and J. R. Bedbrook. 1979. Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucl. Acids Res. 7:1869-1885.
BGN 57 Vol. 21
Heslop-Harrison, J. S., T. Schwarzacher, K. Anamthawat-Jónsson, A. R. Leitch, M. Shi, and I. J. Leitch. 1991. In situ hybridization with automated chromosome denaturation. Technique 3: 109-115.
Leitch, I. J., A. R. Leitch, and J. S. Heslop-Harrison. 1991. Physical mapping of plant DNA sequences by simultaneous in situ hybridization of two differently labelled probes. Genome 34:329-333.
Linde-Laursen, I. 1978. Giemsa C-banding of barley chromosomes 1. Banding pattern polymorphism. Hereditas 88:55-64.
Maluszynska, J., and J. S. Heslop-Harrison. 1991. Localization of tandemly repeated DNA sequences in Arabidopsis thaliana. The Plant Journal 1: 159- 166.
Mukai, Y., T. R. Endo, and B. S. Gill. 1991. Physical mapping of the 18S.26S rRNA multigene family in common wheat: Identification of a new locus. Chromosoma 100:71-78.
Singh, R. H., and T. Tsuchiya. 1977. Morphology, fertility, and transmission in seven monotelotrisomics of barley. Z. Pflanzenzüchtg. 78:327-340.