

The North American Barley Genome Mapping Project (NABGMP) is organized to develop a 10 centiMorgan (cM) map of the barley genome and to use that map to identify quantitative trait loci (QTL) for all of the economically important characteristics of barley. The Project consists of 49 scientists in 26 laboratories in the United States and Canada. It is directed by a project coordinator: Dr. R. A. Nilan (Washington State University), the Canadian co-director: Dr. K. Kasha (University of Guelph), a five-member steering committee, and map construction and economic trait analysis coordinating committees.
Map construction is based on 150 doubled haploid (DH) lines derived from each of two crosses, Morex by Steptoe (six-rowed) and Harrington by TR306 (two-rowed). The parents were selected to represent a broad sample of genetic variation and quantitative trait expression, as well as adequate DNA polymorphism. Morex is the malting industry standard cultivar recommended by the American Malting Barley Association (AMBA). Steptoe is a high yielding, broadly adapted barley with poor malting and feed quality. Morex is a derivative of the Manchurian group while Steptoe is a Coast-type barley. Harrington is the standard two-rowed malting barley in Canada while TR306 is a feed barley with better adaptation, yield, and disease resistance than Harrington.
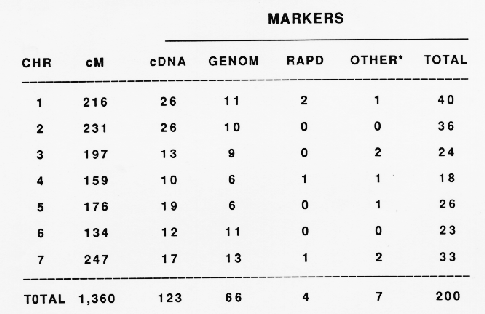
This report presents data on a 200 marker Steptoe/Morex map (Table 1). These include 2 morphological, 5 isozyme, 4 random amplified polymorphic DNA (RAPD), and 189 restriction fragment length polymorphism (RFLP) markers (Table 2). The two morphological markers are rachilla hair length (Srh) and leaf pubescence (Pub). To differentiate these markers from the RFLP markers they are designated with the prefix "m". The isozymes are esterase 5 and 9 (Est5, Est9), phosphogluconate dehydrogenase 2 (Pgd2), alkaline (basic) ,beta-galactosidase (Bg1), and aconitate hydratase 2 (Aco2). The isozyme markers are prefixed with an "i" to differentiate them from the RFLP markers.
The RAPD markers are designated ABR (American Barley Rapd) plus an arbitrary number. The RAPD markers have turned out to be problematic with segregation distortion and reproducibility. They are not recommended for use at this time.
The RFLP markers are from many sources and are either cDNA or genomic DNA probes. The cDNA probes are from the NABGMP libraries (Steptoe leaf; Steptoe malted seed), from Cornell University libraries (barley cDNA, oat cDNA), from the Plant Science Research Institute, Cambridge Laboratory library (Chinese Spring wheat leaf cDNA), and various known function probes obtained from individuals who cloned them (mostly cDNAs). The NABGMP group cNDA probes are designated ABC (American Barley Cdna) plus an arbitrary number. The Cornell probes are designated as in Heun et al. (1991); BCD for barley cDNA and CDO for oat cDNA plus an arbitrary number, except that for markers
where the map location of the marker is different in our (Steptoe/Morex) cross from the Cornell (Proctor/Nudinka) cross, we use a different capital letter designation following the arbitrary number. The Plant Science Research Institute probes are designated PSR plus a number (Chao et al., 1988; Sharp et al., 1988). The known function gene probes are given a locus designation consistent with previous designations or, if none, a symbol is assigned following established guidelines (Barley Genetics Newsletter 11:1-16, 1981). In cases where it was not possible to determine which previously used locus designation corresponded to the locus uncovered by the RFLP probe, a previously unused locus number is used. For example, we mapped five alcohol dehydrogenase loci using the alcohol dehydrogenase cDNA probe (Good et al., 1988), but there are three Adh loci previously identified by isozyme analyses (Søgaard and von Wettstein-Knowles, 1987). Since we were not able to resolve which one was Adhl, 2, or 3, we designated the RFLP detected loci as Adh4, 5, 6, 7, and 8. Similar problems arise with the peroxidase gene loci. Here we have chosen to use the gene symbol Prx for the RFLP identified loci to differentiate them from the more commonly used isozyme symbol Per.
The genomic DNA probes are from a NABGMP barley Pst 1 library (prepared by N. Lapitan), Cornell University barley and wheat libraries (Heun et al., 1991), and a Kansas State University Triticum tauschii library (Gill et al., 1991). The NABGMP probes are designated ABG (American Barley Genomic DNA) plus an arbitrary number. The Cornell University probes are designated BG (barley genomic) or WG (wheat genomic) plus an arbitrary number. The Kansas State University probes are designated ksu followed by a capital letter and an arbitrary number. If a probe detects more than one locus, an additional number or capital letter is added to the designation in an alternating order, i.e. if the designation ends in a letter add a number and if it ends in a number add a capital letter.
The 200 markers mapped to the Steptoe/Morex cross constitute 7 linkage groups spanning 1,360 cM. The distribution of the markers to the seven chromosomes and the genetic length of each are shown in Table 2. Chromosomes 7 and 6 are the longest and shortest, respectively, in recombination distance. Chromosomes 1 and 4 have the most and the least number of markers mapped, respectively. The centromere location has been initiated by mapping markers to chromosome arms using telosomic barley-wheat addition lines (Shepherd and Islam, 1987). We suspect that in many cases we have not yet reached the ends of the chromosomes. Putative telomeric markers have been mapped to the short arm of chromosomes 1 and 2 (A. Kilian, unpublished). Other chromosome arm telomeres remain to be mapped. The maps are presented in Fig. 1.
In order to facilitate communications, we have developed an electronic bulletin board for the barley genome mapping effort. This bulletin board is located at Washington State University, the address is "WSUVMS1.CSC.WSU.EDU", the user name is "BARMAPBB" and the current password is "STEPTOE". Eventually the bulletin board will contain up to date barley mapping information, standard techniques, and messages of general interest.
Outside users will be able to leave messages on the bulletin board and download data to their personal computers.
The NABGMP group is an open organization welcoming any participants who wish to contribute to its goals. Interest individuals should contact the project coordinators Dr. R. A. Nilan, Depts. Crop & Soil Sciences and Genetics & Cell Biology, Washington State University, Pullman, WA 99164-6420, USA, FAX: 509-335-8674, E-mail: NILANRA@WSUVM1.BITNET or Dr. Ken Kasha, Dept. Crop Science, University of Guelph, Guelph, Ontario, Canada, N1G 2W1, FAX: 519-763-8933. Seeds from the parents and doubled haploid lines of the Steptoe/Morex cross are available from Dr. Pat Hayes, Crop Science Dept., Oregon State University, Corvallis, OR 97331, USA, FAX: 503-737-1589, Email: HAYES@CCMAIL.ORST.EDU. Seeds from the parents and doubled haploid lines of the Harrington/TR306 cross are available from Dr. K. J. Kasha. The probes that are property of NABGMP and others that we have perrnission to distribute are available from Dr. Andy Kleinhofs, Depts. Crop & Soil Sciences and Genetics & Cell Biology, Washington State University, Pullman, WA 99164-6420, USA, FAX: 1-509-335-8674, E-Mail: COLECO@WSUVMS1.BITNET.
References:
Choa, S., P. J. Sharp, and M. D. Gale. 1988. A linkage map of wheat homoeologous group chromosomes using RFLP markers. In: T. E. Miller and R. M. D. Koebner, eds., Proc. Seventh International Wheat Genetics Symposium, Vol. 1 (Cambridge, England, 1988). Institute of Plant Science Research, Cambridge. pp. 493-498.
Gill, K., E. Lubbers, B. S. Gill, W. J. Raupp, and T. S. Cox. 1991. A genetic linkage map of Triticum tauschii (DD) and its relationship to the D genome of red wheat (AABBDD). Genome 34:362-374.
Good, A. G., L. E. Pelcher, and W. L. Crosby. 1988. Nucleotide sequence of a complete barley alcohol dehydrogenase 1 cDNA. Nucleic Acids Research 16:7182.
Heun, M., A. E. Kennedy, J. A. Anderson, N. L. V. Lapitan, M. E. Sorrells, and S. D. Tanksley. 1991. Construction of a restriction fragment length polymorphism map for barley (Hordeum vulgare). Genome 34:437-447.
Lander, E. S., et al. 1987. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations, Genomics 1:185-199.
BGN 48 Vol.21
Liu, B. H., and S. J. Knapp. 1990. GMENDEL: A program for Mendelian segregation and linkage analysis of individual or multiple progeny populations using log-likelihood ratios. J. Hered. 81:407.
Proctor, L., A. Rafalski, S. Tingey, S. Lincoln, and E. Lander. 1987. Mapmaker macintosh V1.0 for the construction of genetic linkage maps. Copyright 1987 WIBR; copyright 1990 E. I. duPont de Nemours and Co.
Sharp, P. J., S. Desai, S. Chao, and M. D. Gale. 1988. Isolation and use of a set of fourteen RFLP probes for the identification of each homoeologous chromosome arm in the Triticeae. In: T. E. Miller and R. M. D. Koebner, eds., Proc. Seventh International Wheat Genetics Symposium, Vol. 1 (Cambridge, England, 1988). Institute of Plant Science Research, Cambridge, pp. 639-646.
Shepherd, K. W., and A. K. M. R. Islam. 1987. Cytogenetic manipulation of barley chromosomes in a wheat background. In: S. Yasuda and T. Konishi, eds. Barley Genetics V. Proc. Fifth International Barley Genetics Symposium, (Okayama, Japan, 1987) pp. 375-387. Sanyo Press Co., Okayama.
Søgaard, B., and P. von Wettstein-Knowles. 1987. Barley: genes and chromosomes. Carlsberg Res. Commun. 52:123-196.