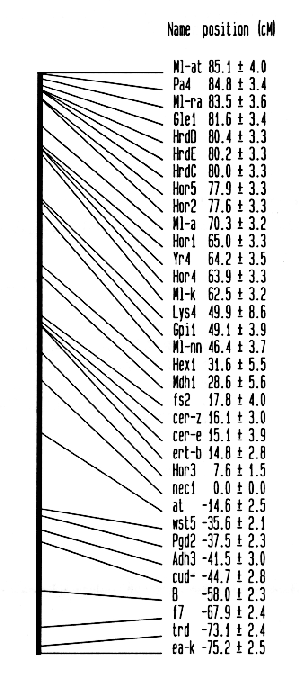
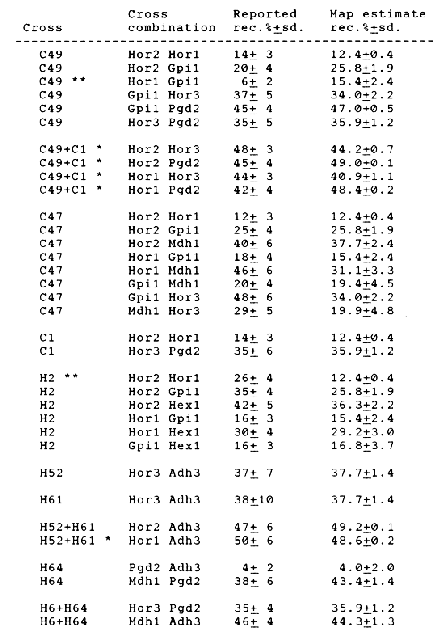
Based on crosses between lines drawn from Composite Cross 34 and landraces from Iran (the C series) or between lines of H. spontaneum and H. vulgare cultivar 'Clipper' (the H Series), four new isoenzyme loci, Gpil (Glucosephosphate isomerase), Hex1 (Hexokinase), Mdh1 (Malate dehydrogenase), and Adh3 (Alcohol dehydrogenase) can be mapped on barley chromosome 5. The linkage data were reported by Brown et al. (1989). The data are shown in Table 1. The linkage map estimation procedure reported by Jensen and Jorgensen (1975) has been applied to previously published data and to the data given in Table 1. The data marked with a star (*) were excluded as they are confounded with the other data. Further, they have very little information. The mapping procedure estimated the map shown in Fig. 1. Two recombination frequencies did not fit the map and were discarded by the procedure, they are marked with two stars (**).
References:
Brown, A. H. D., G. J. Lawrence, M. Jenkin, J. Douglass, and E. Gregory. 1989. Linkage drag in backcross breeding in barley. J. Hered. 88: 234 - 239.
Jensen, J., and J. H. Jorgensen. 1975. The barley chromosome 5 linkage map. I. Literature survey and map estimation procedure. Hereditas 80:5-16.