Chlorina mutants of barley are useful linkage stocks, since they are easily identified by yellow or light green leaves at the seedling stage. The chlorina characteristics are governed by different genes at 12 loci, f to f11 and fc. Among them, the specific locations of the f2, f3, f6 and f11 loci are not determined yet. The chlorina mutant carrying the f2 gene is obviously different from the normal green type by yellow color until the heading stage, even when it is grown under any condition. Tsuchiya and Singh (1982) located the f2 locus on the short arm of chromosome 3 by means of telotrisomic analysis, whereas Tsuchiya et al. (1984) revised the short arm to the long one by acrotrisomic analysis. Therefore, the f2 locus should be considered to be located on the long arm of chromosome 3.
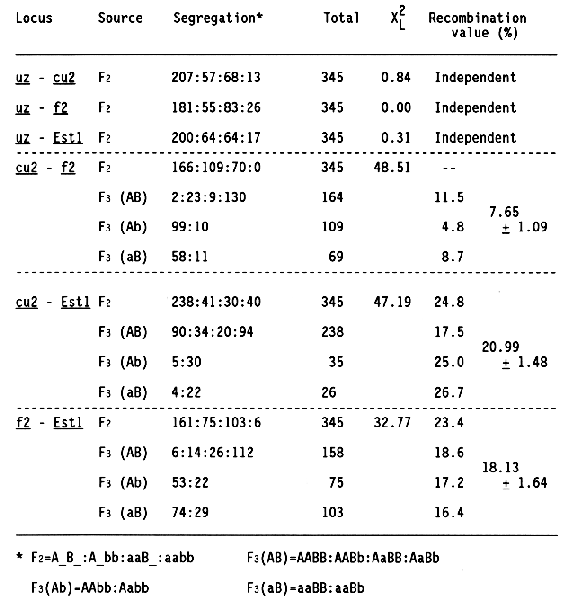
For linkage analysis, a total of 345 F2 individuals and their 342 F3 progenies derived from a cross combination between Chlorina 2 (BGS 117) and Choshiro-hen (BGS 114) were used. At the seedling stage, segregation of F2 individuals and that of F3 plants within each progeny were examined for the chlorina seedling (F2), together with uzu type (uz), curly growth (cu2), and 4 esterase isozymes (Estl). These characters are controlled by different genes which are located on the long arm of chromosome 3. The results are shown in Table 1.
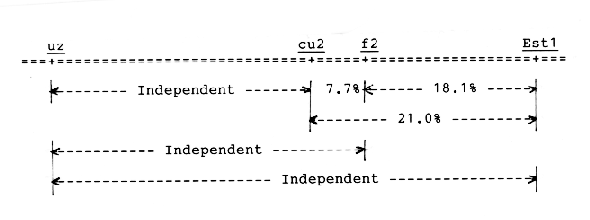
The uz locus is independent of the other three loci, cu2, f2, and Est1. The cu2 and f2 loci are tightly linked with each other, and they are also linked with the Est1, which is located at the terminal of the long arm of chromosome 3 (Konishi and Matsuura, 1987). From the recombination values indicated in Table 1, the four loci are arranged in the order of uz, cu2, f2, and Est1 from the proximal to the terminal of the long arm of chromosome 3, as shown in Fig. 1.
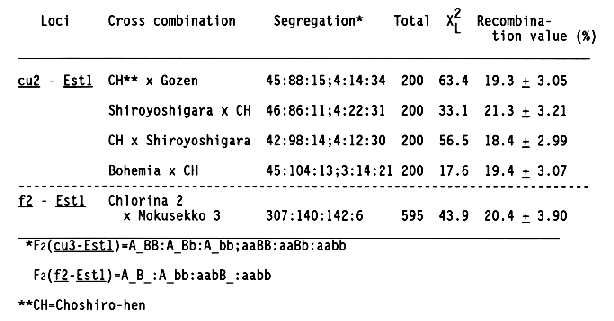
The order of two loci, cu2 and f2, was determined by the method of three-point test (Smith, 1947) because these loci were closely linked with each other. Because segregation of curly growth and chlorina seedling in F2 and F3 generations of this cross combination were somewhat distorted, the recombination values between cu2 and Est1 and between f2 and Est1 were estimated using data from other cross combinations which showed the normal segregation (Table 2). However, these recombination values agreed with those in Table 1.
References:
Konishi, T., and S. Matsuura. 1987. Linkage analysis of Est4 locus for esterase isozyme-4 in barley. BGN 17:68-70.
Smith, L. 1947. Simple method for establishing the three-point order of genes from F3 data. Jour. Amer. Soc. Agron. 37:353-355.
Tsuchiya, T., and R. J. Singh. 1982. Chromosome mapping in barley by means of telotrisomic analysis. Theor. Appl. Genet. 61:201-208.
Tsuchiya, T., R. J. Singh, Azizeh Shahla and A. Hang. 1984. Acrotrisomic analysis in linkage mapping in barley (Hordeum vulgare L.). Theor. Appl. Genet. 68:433-439.