

The M1-a disease reaction locus is one of a set of loci which define the response of barley to Erysiphe graminis f. sp. hordei, the causal agent of the powdery mildew disease. In experiments to define the genetic fine structure of this locus, F3 families with susceptible variants were recovered, from a cross between two resistant lines, at an unusually high frequency (Wise and Ellingboe, 1985). Roughly 0.6% of the F3 families contained susceptible members; a frequency 10-100 times higher than observed among F3 families of other crosses. Susceptible variants were subsequently identified in an F2 population at a frequency of 1.67%. This apparent high frequency of recombination between alleles at the M1-a locus was only observed in progeny from crosses between barley lines CI-16151 (female) and CI-16155. No susceptible variants were found among progeny derived from the reciprocal cross. Furthermore, susceptible variants often reverted to resistance in subsequent generations. We have repeated the experiments of Wise and Ellingboe (1985) in an attempt to confirm their observation and to extend our knowledge of the nature of this genetic instability.
A high frequency of apparent recombination and/or mutation is often associated with the action of transposable elements (Nevers et al., 1986), or with paramutation (Brink, 1973). To study the molecular basis of the genetic instabiity observed in the F2 and F3 progeny, it is convenient to monitor unstable events at a well-characterized gene. Therefore, in addition to the M1-a locus, we screened for variants at the alcohol dehydrogenase isozyme 1 (ADH1) and waxy genes, because rapid phenotypic screens exist and cloned genes are available for use as probes of gene structure. We describe attempts to recover variants at the M1-a, waxy or ADH1 genes in F2 (CI-16151 x CI-16155) progeny.
Genetic Stocks: Barley lines CI-16151 (M1-a6), CI-16155 (M1-a13), CI-16138 (m1-a), CI-16137 (M1-a), CI-16141 (M1-h), and CI-16139 (M1-9) were obtained from R. Wise [see Moseman (1972) for a description of the lines]. The waxy mutant (gamma, field 81) was a gift from Drs. A. Kleinhofs and R. Nilan (Washington State University, Pullman, WA) and the ADH1- mutant, CPI-96981-5, was obtained from Dr. Edwards (designated M9 in Harberd and Edwards, 1982). Erysiphe graminis f. sp. hordei, race CR3 was obtained from R. Wise. Crosses between CI-16151 and CI-16155 were made in a growth chamber using standard methods (Starling, 1980). F1 seeds from the various crosses were grown to maturity in a growth chamber isolated from other genotypes, to produce the F2 populations utilized for these studies. Seed from each F1 plant was harvested and analyzed separately.
Screen for Powdery Mildew Resistance: In initial experiments, seeds were out in half and the embryo-half was planted directly into jiffypots. However the number of normal, healthy seedlings recovered was low. Using the method described below roughly 90% of the seeds gave rise to normal seedlings. Seeds were cut in half and the embryo halves were germinated on agar plates containing mineral nutrients (Somerville and Ogren, (1982), in the dark at room temperature for 2 days. The seedlings were transferred to a light rack for one day, and were then transplanted to 3-inch "jiffypots" containing a 1:1:1 mix of vermiculite, perlite and sphagnum. The seedlings were grown for 1 week in growth chambers with 100-150 uE PAR M-2S-1 (mixed cool white fluorescent and incandescent lights), with a 16 h photoperiod, and day and night temperatures of 22°C and 18°C respectively. Seedlings were then transferred to isolated growth chambers and inoculated with race CR3. The conditions for disease development were 50 uE PAR M-2S-1 (mixed cool white and incandescent lights), with a 15 h photoperiod, and day and night temperatures of 180C and 160C respectively (Masri and Ellingboe, 1966). Disease reaction scores (Moseman, 1972) were determined 2 weeks following inoculation. Lines CI-16138, CI-16137, CI-16141, CI-16151 and CI-16139, each of which carries a different resistance allele, were tested at the same time to verify the identity of race CR3.
Waxy Phenotype Screen: Endosperm quarters from each seed were tested for the waxy phenotype by staining with iodine (Ho et al., 1980). Starch lacking amylose, the waxy- phenotype, stains a red-brown color, while the waxy+ phenotype gives a black color in this assay. Endosperm quarters were incubated in a 96-well microtiter plate in 200 ul of KI-I2 solution (8.67 mM KI, 0.56 mM I2 in 0.04 N HC1) for 60 min with shaking (100 rpm) at room temperature. The seed pieces were then scored visually for black color. The waxy mutant was included as a negative control and the two parental lines served as positive controls.
ADH1 Phenotype Screen: Endosperm quarters were incubated in 24-well plates in 400 ul of freshly prepared ADH activity stain (25 mM Tris-Cl pH 8, 2.5 mM NADF+, 2.5 mM Nitro Blue Tetrazolium 25% ethanol) with shaking for 5 min. The reaction was stopped by the addition of 1 ml 0.01 N HC1, and the endosperm piece was observed at 25× magnification using a stereomicroscope. In the aleurone layer, ADH1 is the only ADH isozyme expressed constitutively at significant levels (Hanson et al., 1984). A dark purple ring of cells around the endosperm is indicative of wild-type levels of ADH1 activity (Harberd and Edwards, 1982). the ADH1- mutant, CPI-96981-5, was included as a negative control and the parental lines were included as positive controls. ADH Isozyme Analysis: Barley roots were harvested from 3-week-old seedlings grown hydroponically. Isozymes of ADH were induced by bubbling N2 through the hydroponic medium for 3 days prior to harvest. Roots were ground in 0.15 M Tris-Cl pH 8, 10 mM dithiothreitol (2 ml/g roots) at 4°C; the extract was clarified by centrifugation; and 45 ul of supernatant,was analyzed on native polyacrylamide gels. Extracts from endosperm quarters from dry seeds were prepared and analyzed as described for root tissue. Electrophoresis and staining for ADH activity was carried out as described by Hanson et al. (1984).
Hordein Characterization: Lines CI-16151 and CI-16155 have distinct hordein banding patterns which are useful for checking the genotypes of these lines (Wise and Ellingboe, 1985). Hordeins were isolated from half a seed and separated by polyacrylamide gel electrophoresis under denaturing conditions according to the protocol of Doll and Anderson (1981).
Southern Analysis: DNA was isolated from 2-4 week old seedlings by a miniprep method (Dellaporta et al., 1983). Southern blots were prepared using standard procedures (Maniatis et al., 1982). In brief, DNA was digested with various restriction endonucleases, separated by electrophoresis in 0.9% agarose gels and transferred to nylon membranes (Zetaprobe, Biorad) by the alkaline transfer method (Reed and Mann, 1985). Filters were probed with the 6-kbp EcoR1 insert of pM9-15.1, which contains most of the coding sequence and about 4-kbp of 5' noncoding region of the ADH1 barley gene (Trick et al., in press). The 6-kbp insert was purified from 1% low melting agarose gels on NACS columns (Prepacs, BRL), and labelled by the random primer method (Feinberg and Vogelstein, 1983) to a specific activity greater than 108 cpm/ug (Oligolabeling kit, Pharmacia). Hybridization was carried out overnight at 42°C in 50% formamide, 5× SSC, 5× Denhart's solution, 1% sodium dodecyl sulfate, 500 ug salmon sperm DNA/ml and 10% Dextran Sulfate. The filters were then washed at increasing stringencies culminating with 0.1× SSC, 0.1% sodium dodecyl sulfate at 68°C. Filters were dried and exposed for 4-6 days to Kodak XAR-5 x-ray film at -80°C using intensifying screens.
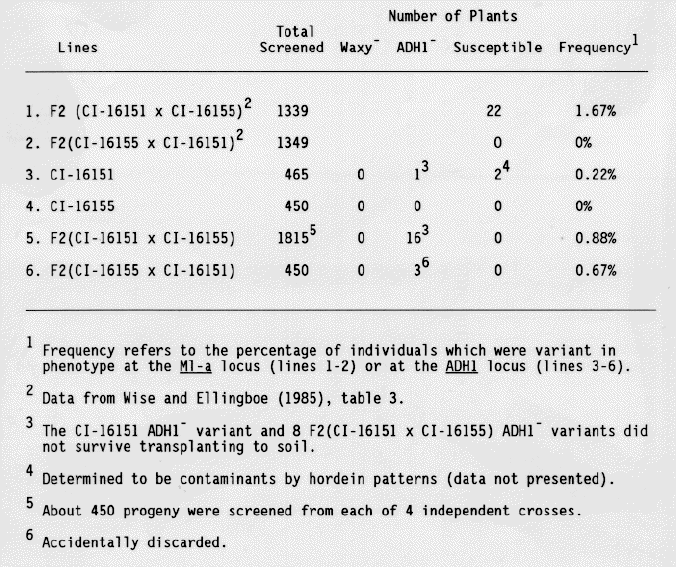
F2 seeds from crosses between CI-16151 and CI-16155 were screened for ADH1- and waxy- variants as well as for variants susceptible to E. graminis, race CR3. As shown in Table 1, no waxy or susceptible variants were identified. Two susceptible variants were recovered from field grown stocks of CI-16151, however, these were shown to be contaminants based on their hordein phenotypes. All subsequent tests were done using growth chamber-grown stocks. Twenty ADH1- seeds were identified. The recovery of ADH- variants was only slightly less frequent than the recovery of susceptible variants as described in previous work (Wise and Ellingboe, 1985).
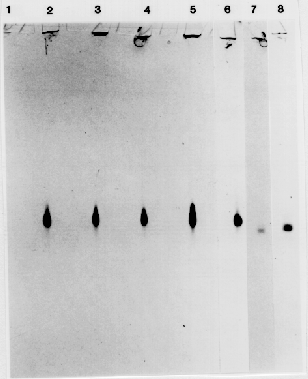
Eight of the 16 ADH1 variants from the F2 (CI-16151 × CI-16155) progeny survived to produce seed. About 16 F3 seeds from each of the 8 putative mutants were tested for ADH1 activity in the aleurone. All progeny exhibited wild-type levels of ADH1 activity. Seeds from the ADH1- control plants, grown under the same conditions, displayed a higher than normal, degree of staining. Therefore, we investigated whether ADH2 or ADH3 activity had been induced during development of these seeds, thereby creating the false impression that the variants had ADH1 activity. The ADH isozymes were separated by native polyacrylamide gel electrophoresis and identified using the ADH activity stain. Progeny from the F3 lines displayed significant levels of ADH1 activity in both anaerobically- and aerobically- grown (data not presented), and in dry, aerobic seeds (Fig. 1), confirming the results of the aleurone stain.
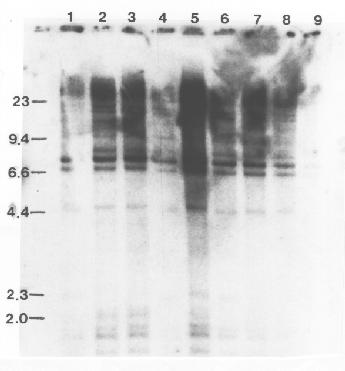
Structural alterations at the ADH1 gene, such as insertions and deletions characteristic of transposon-induced mutations, should be observed as changes between the parents and progeny in the banding pattern of genomic DNA following digestion with restruction endonucleases and hybridization with the cloned ADH1 gene. No restriction fragment length polymorphisms were observed among the variants and the parental lines (Fig. 2). The blots in Fig. 2 were also overexposed, so that minor bands and bands in underloaded lanes could be observed, and no polymorphisms were
References:
Brink, R. A. 1973. Dellaporta, S. L., J. minipreparation: Paramutation. Ann. Rev. Genet. 7:129-152.
Wood and J. B. Hicks. 1983. A plant DNA version II. Plant Mol. Bio. Rep. 1:19-21.
Feinberg, A. P. and B. Vogelstrin. 1983. A technique for radio- labeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132:6-13 [see addendum in Anal. Biochem. 137-266-267 (1984).
Doll, H. and B. Anderson. 1981. Preparation of barley storage protein, Hordein, for analytical sodium dodecyl sulfate- polyacrylamide gel elelctrophoresis. Anal. Biochem. 115, 61-66.
Harberd, N. P. and K. J. R. Edwards. 1982. A mutational analysis of the alcohol dehydrogenase system in barley. Heredity 48:187-195.
Hanson, A. D.j J. V. Jacobson and J. A. Zwar. 1984. Regulated expression of three alcohol dehydrogenase genes in barley aleurone layers. Plant Physiol 75:573-581.
Ho, D. T. H., S. C. Shih, and A. KleinhofB. 1980. Screening for barley mutants with altered hormone sensitivity in their aleurone layers. Plant Physiol. 66:153-157.
Maniathis, T., E. F. Fritsch and J. Sambrook. 1982. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
Masri, S. H. and A. H. Ellingboe. 1966. Primary infection of wheat and barley by Erysiphe graminis. Phytopathol. 56, 389-395.
Moseman, J. G. 1972. Isogenic barley lines for reaction to Erysiphe graminis,f.sp. hordei. Crop Sci. 12:681-682.
Nevers, P., N. S. Shepherd and H. Saedler. 1986. Plant trans- posable elements. Adv. Bot. Res. 12:104-203.
Reed, K. C. and D. A. Mann. 1985. Rapid transfer of DNA from agarose gels to nylon membranes. Nucl. Acid. Res. 13:7207-7221.
Somerville, C. R. and W. L. Ogren. 1982. Isolation of photo-respiration mutants in Arabidopsis thaliana. In: Methods in Chloroplast Biology. Edelman, M., R. B. Hallick and N. H. Chua (eds.). Elsevier, Amsterdam. pp. 129-138.
Starling, T. M. 1980. Barley. In: Hybridization of Crop Plants. Fehr, W. R. and H. H. Hadley (eds). American Society of Agronomy Publishers, Madison, WI. pp. 189 202.
Trick, M., E. S. Dennis, K. J. R. Edwards and W. J. Peacock. Molecular analysis of the alcohol dehydrogenase (ADH) gene family of barley. In press.
Wise, R. P. and A. H. Ellingboe. 1985. Fine structure and instability at the M1-a locus in barley. Genetics 111:113-130.