Bikram S Gill, Wheat Genetics Resource Center, Plant Pathology Department, Kansas State University, Manhattan, KS 66506-5502, USA.
At a recent meeting of ITMI in Winnipeg, Canada (1-4 June, 2002), cereal workers discussed the concept of IGROW to lead wheat genomic and improvement effort for the next 10 years. Dubbed as the IGROW 2010 project and similar to the Arabidopsis 2010 project to elucidate a functioning plant model, the vision of IGROW is rather modest:
The IGROW challenge is to coördinate and provide direction to diverse wheat research in the following ten areas:
I propose that we identify our best scientists who will coördinate and lead research in the above mentioned areas. I envision community resources and a research effort in each of the above areas. I propose that Annual Wheat Newsletter publish research as short notes in all of the above areas after peer review by respective focus groups. It will be a major change in the mission of Annual Wheat Newsletter to become an instrument that will provide leadership at this critical juncture.
Steering committee: Rudi Appels (Australia) and Bikram Gill (USA), lead coordinators; Olin Anderson (USDA-ARS, USA); Boulos Chalhoub (URGV-INRA, France); Jun Yu (BGI, China); Yasumari Ogihara (Japan); Beat Keller (Switzerland); and Graham Moore (UK). The committee will expand as additional countries join the project.
One of the first challenges of IGROW is selective sequencing of the bread wheat genome to access most of the genes in the wheat plant. Some arguments that can be advanced for wheat genome sequencing are as follows.
Concepts and aims. The IGROW project will provide an international infrastructure to integrate existing wheat sequencing projects and a mechanism to leverage new funds for the sequencing of the gene-rich regions from bread wheat. The focus on gene-rich regions aims to capture structural information on agronomically important genes which are generally crop-specific.
Background. Cultivated wheats belong to three ploidy levels, diploid (einkorn), tetraploid (emmer or durum), and hexaploid (dinkel, common, or bread). It is the hexaploid or bread wheat (genome size 16 billion bp) that is of overwhelming economic importance. Wheat is grown on more areas of land than any other crop plant and is a staple food of 40 % of the world's population. It provides for 20 % of the calories consumed. Wheat originated in the Fertile Crescent, an arc of land stretching from the Persian Gulf, north to Turkey, and south again to Egypt. The fertile soil watered by rivers like the Euphrates and the Jordan enabled humans to invent farming, cities, trade, and writing. The Sumerians formed the first empire in 4000 BC followed by Babylonia, Assyria, Mesopotamia, and Arabia. The development of wheat as a food source is deeply linked into this history of modern civilization. The present proposal will identify the genes that were critical in this development. The large-scale, genome-sequencing proposed in this project interfaces with studies worldwide to identify disease-resistance and abiotic resistance genes and quantitative trait loci defining agronomic and quality traits and will, therefore, include genes that helped establish human civilization.
Chinese Spring is a bread wheat land race from China that has been used as a genetic model for over half a century by the international wheat genetics community. A vast array of cytogenetic stocks and genetic resources have been developed for trait discovery. During the past few years, several genomics projects on bread wheat have been established in different countries and include the sequencing of over 200,000 ESTs from over 50 different wheat tissues and the large-scale mapping of EST and BAC clones. These wheat genome projects and resources will constitute the starting material for large-scale genome sequencing. Gene discovery findings through this large-scale genomic sequencing will drive the continued adaptation of wheat with respect to both agronomic and quality traits. New grain storage protein genes discovered through functional genomics will, for example, provide for novel approaches to altering quality traits in breeding programs. The creation of specific food products for human consumption and health, and maintaining crop production in difficult environments will depend on utilizing novel genes revealed by large-scale sequencing.
The polyploid nature of wheat has allowed nature to 'experiment' with its genetic material in a way not possible in other major crops or well-studied systems such as Arabidopsis and rice. New versions of genes have, for example, been discovered in the analysis of starch biosynthesis.
Molecular and cytogenetic mapping of ESTs in chromosome bins using deletion stocks has revealed that the wheat genome is sharply partitioned into gene-poor and gene-rich components. The gene-rich regions are accessible either by EST probing of large DNA (BAC) libraries or generalized genome-fractionation methods for a large genome-sequencing project. The increased efficiency and lower cost of large-scale genome sequencing makes it feasible to plan a project as large as sequencing the gene-rich regions of the wheat genome.
Proposal. In view of the large size of the genome, it is proposed that IGROW will focus on the gene-rich regions of the wheat genome. Three phases can be defined:
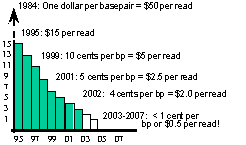 The estimated budgets
for the phases, based on the Beijing rice sequencing project (published
in April issue of Science, 2002) are identified in Fig. 1 (to
the left) and Fig. 2 (below). Note the reduction in the cost of
sequencing over time. The IGROW estimate for sequencing the gene-rich
portion of the wheat genome is $100 M (Fig. 2, below)
The estimated budgets
for the phases, based on the Beijing rice sequencing project (published
in April issue of Science, 2002) are identified in Fig. 1 (to
the left) and Fig. 2 (below). Note the reduction in the cost of
sequencing over time. The IGROW estimate for sequencing the gene-rich
portion of the wheat genome is $100 M (Fig. 2, below)
Integration of other sequencing activities. The extensive sequencing of expressed genes (cDNAs) in wheat, especially in Japan, the U.S., and Europe, with a focus on full-length cDNAs, is essential for the annotation of the genes in the genomic-sequencing work. This also will provide the basis for capturing the functional assignment activity for similar genes in other organisms such as Arabidopsis and rice. Many agronomic and quality related genes will have functions unique to wheat, and the genome-sequencing project will form the basis for investigating the functional attributes of these genes in more detail.
A major resource that makes the present proposal feasible is
the extensive molecular genetic maps that have 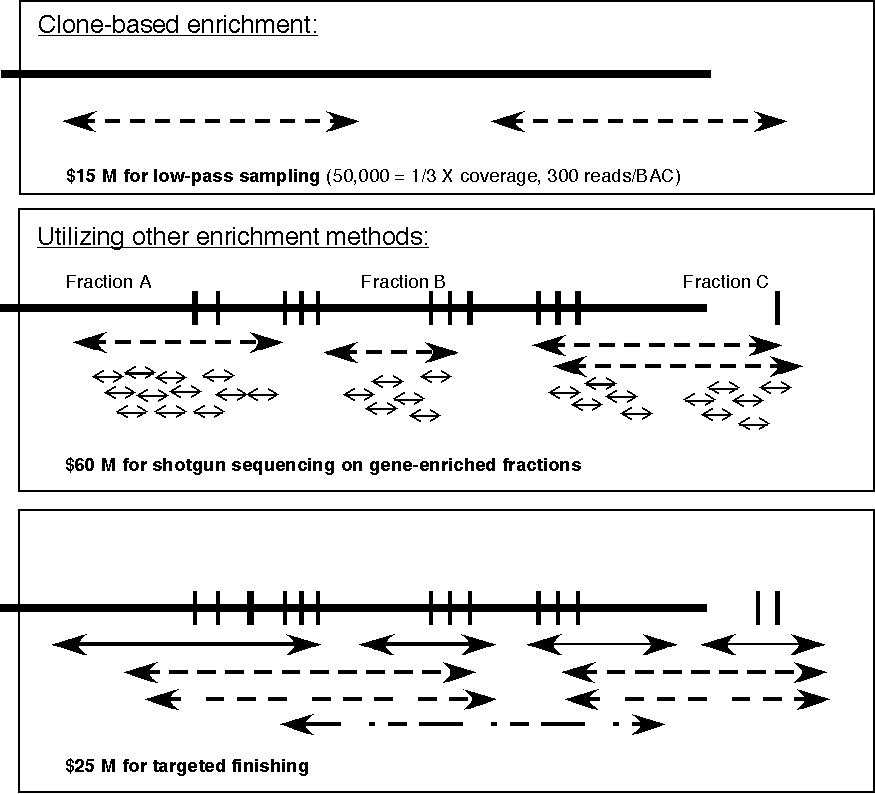 been
established for wheat. In addition, the assignment of the location
of over 10,000 ESTs within the molecular genetic map using chromosome
deletion stocks, currently funded by the National Science Foundation
(NSF, USA), is starting the process of identifying candidate genes
associated with QTLs of importance in wheat. These resources provide
the basis of positioning and orienting the large-scale sequencing
effort. Large-scale sequencing of wheat ESTs in Japan, especially
full-length cDNA sequencing will be crucial for interpreting the
sequence structure of the genome.
been
established for wheat. In addition, the assignment of the location
of over 10,000 ESTs within the molecular genetic map using chromosome
deletion stocks, currently funded by the National Science Foundation
(NSF, USA), is starting the process of identifying candidate genes
associated with QTLs of importance in wheat. These resources provide
the basis of positioning and orienting the large-scale sequencing
effort. Large-scale sequencing of wheat ESTs in Japan, especially
full-length cDNA sequencing will be crucial for interpreting the
sequence structure of the genome.
The major resources for the wheat genome project established in Europe include the BAC library in the cultivar Renan available from the French Genoplante Consortium and the BBSRC/INRA BAC library from Chinese Spring; the large-scale, hexaploid wheat EST sequencing in France (Genoplante Consortium); and a project to anchor 10,000 ESTs on the Renan BAC library and the sequencing of 2-3 Mb segments of the genome.
A significant resource that will complement this project is the current investment by the NSF (USA) in establishing a complete representation of the D genome of wheat (Ae. tauschii) by BAC clones. The analysis of this resource will be particularly significant in phase 3 of the project proposed, where the detailed structure of specific regions will be greatly aided by the availability of homoeologous genomic clones from a diploid donor of one of the wheat genomes. Similarly, the detailed analyses of the T. monococcum BAC library clones by Keller and colleagues (Switzerland) and Dubcovsky and colleagues (USA) also will provide a key resource for refining analyses of specific regions of the wheat genome.
Plan of action. This preliminary proposal has been developed for the benefit of individual scientists and organizations for obtaining funding in their respective countries for participating in the IGROW project. All sequence data will be in the public domain. You may contact Rudi Appels (rappels@agric.wa.gov.au) or Bikram Gill (bsg@ksu.edu) for further information.