|
|
[ Download Map Data ]
[ Download Feature Correspondence Data ]
|
|
|
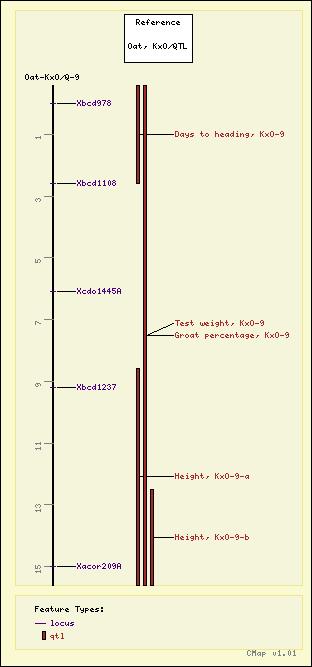
Oat - Oat, KxO/QTL - Oat-KxO/Q-9
(Click headers to resort) |
Comparative Maps |
| Feature |
Type |
Position () |
Map |
Feature |
Type |
Position |
Evidence |
Actions |
|
Xacor209A
|
locus
|
15
|
Oat - Oat, KxO - Oat-KxO-9
|
Xacor209A
|
locus
|
15 ()
|
Automated name-based
|
View Maps
|
|
Oat - Oat-2001-KxO-QTL - Oat-2001-KxO-QTL_9
|
Xacor209A
|
locus
|
10 (cM)
|
Automated name-based
|
View Maps
|
|
Xbcd978
|
locus
|
0
|
Oat - Oat-2003-OglexMAM17-5 - Oat-2003-OglexMAM17-5_13
|
Xbcd978
|
locus
|
63 (cM)
|
Automated name-based
|
View Maps
|
|
Oat - Oat-2003-OxM-QTL1 - Oat-2003-OxM-QTL1_20
|
Xbcd978
|
locus
|
63 (cM)
|
Automated name-based
|
View Maps
|
|
Oat - Oat-2003-OglexMAM17-5 - Oat-2003-OglexMAM17-5_27
|
Xbcd978
|
locus
|
63 (cM)
|
Automated name-based
|
View Maps
|
|
Oat - Oat-2003-OxM-QTL1 - Oat-2003-OxM-QTL1_6
|
Xbcd978
|
locus
|
63.2 (cM)
|
Automated name-based
|
View Maps
|
|
Oat - Oat, KxO - Oat-KxO-9
|
Xbcd978
|
locus
|
0 ()
|
Automated name-based
|
View Maps
|
|
Oat - Oat-2003-OglexMAM17-5 - Oat-2003-OglexMAM17-5_6
|
Xbcd978
|
locus
|
63 (cM)
|
Automated name-based
|
View Maps
|
|
Diploid oat - Diploid Oat, Cornell2 - Avena-Sorrells-B
|
Xbcd978
|
locus
|
17.4 ()
|
Automated name-based
|
View Maps
|
|
Oat - Oat-2003-OglexMAM17-5 - Oat-2003-OglexMAM17-5_20
|
Xbcd978
|
locus
|
63 (cM)
|
Automated name-based
|
View Maps
|
|
Oat - Oat-2003-OglexMAM17-5 - Oat-2003-OglexMAM17-5_6
|
Xbcd978
|
locus
|
63 (cM)
|
Automated name-based
|
View Maps
|
|
Oat - Oat-2001-KxO-QTL - Oat-2001-KxO-QTL_9
|
Xbcd978
|
locus
|
0 (cM)
|
Automated name-based
|
View Maps
|
|
Oat - Oat-2004-OxM-QTL4 - Oat-2004-OxM-QTL4_6
|
Xbcd978
|
locus
|
63.2 (cM)
|
Automated name-based
|
View Maps
|
|
Xbcd1108
|
locus
|
2.6
|
Oat - Oat, KxO - Oat-KxO-9
|
Xbcd1108
|
locus
|
2.6 ()
|
Automated name-based
|
View Maps
|
|
Xbcd1237
|
locus
|
9.2
|
Oat - Oat-2003-OxM-QTL1 - Oat-2003-OxM-QTL1_20
|
Xbcd1237
|
locus
|
66 (cM)
|
Automated name-based
|
View Maps
|
|
Oat - Oat-2003-OglexMAM17-5 - Oat-2003-OglexMAM17-5_20
|
Xbcd1237
|
locus
|
66 (cM)
|
Automated name-based
|
View Maps
|
|
Oat - Oat-2004-OxM-QTL4 - Oat-2004-OxM-QTL4_6
|
Xbcd1237
|
locus
|
65.9 (cM)
|
Automated name-based
|
View Maps
|
|
Diploid oat - Diploid Oat, Cornell2 - Avena-Sorrells-B
|
Xbcd1237
|
locus
|
15.4 ()
|
Automated name-based
|
View Maps
|
|
Oat - Oat-2003-OglexMAM17-5 - Oat-2003-OglexMAM17-5_27
|
Xbcd1237
|
locus
|
66 (cM)
|
Automated name-based
|
View Maps
|
|
Oat - Oat, KxO - Oat-KxO-9
|
Xbcd1237
|
locus
|
9.2 ()
|
Automated name-based
|
View Maps
|
|
Oat - Oat-2003-OglexMAM17-5 - Oat-2003-OglexMAM17-5_6
|
Xbcd1237
|
locus
|
66 (cM)
|
Automated name-based
|
View Maps
|
|
Oat - Oat-2003-OxM-QTL1 - Oat-2003-OxM-QTL1_6
|
Xbcd1237
|
locus
|
66 (cM)
|
Automated name-based
|
View Maps
|
|
Oat - Oat-2003-OglexMAM17-5 - Oat-2003-OglexMAM17-5_6
|
Xbcd1237
|
locus
|
66 (cM)
|
Automated name-based
|
View Maps
|
|
Oat - Oat-2003-OglexMAM17-5 - Oat-2003-OglexMAM17-5_13
|
Xbcd1237
|
locus
|
66 (cM)
|
Automated name-based
|
View Maps
|
|
Xcdo1445A
|
locus
|
6.1
|
Oat - Oat, KxO - Oat-KxO-9
|
Xcdo1445A
|
locus
|
6.1 ()
|
Automated name-based
|
View Maps
|
|
Test weight, KxO-9
|
qtl
|
-0.6 - 15.6
|
No other positions |
|
Groat percentage, KxO-9
|
qtl
|
-0.6 - 15.6
|
No other positions |
|
Days to heading, KxO-9
|
qtl
|
-0.6 - 2.6
|
No other positions |
|
Height, KxO-9-a
|
qtl
|
8.6 - 15.6
|
No other positions |
|
Height, KxO-9-b
|
qtl
|
12.5 - 15.6
|
No other positions |