GrainGenes
A Database for Triticeae and Avena
 |
|
*Bookmarks for this page will fail after this session expires.
Use the "Save Link" button to create a permanent link
|
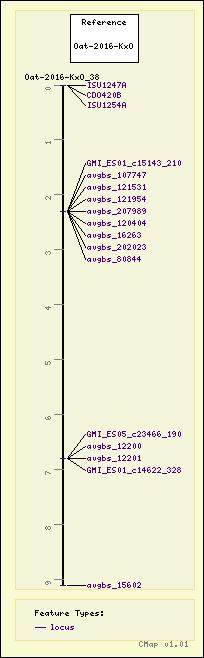
Map Details
| Map Type: | Genetic [ View Map Type Info ] | ||||
|---|---|---|---|---|---|
| Map Set Name: | Oat - Oat-2016-KxO [ View Map Set Info ] | ||||
| Map Name: | Oat-2016-KxO_38 | ||||
| Map Start: | 0 | ||||
| Map Stop: | 9.1 | ||||
| Features by Type: |
|
Map Features
| [ Download Map Data ]
[ Download Feature Correspondence Data ] |
||||||||
| Oat - Oat-2016-KxO - Oat-2016-KxO_38 (Click headers to resort) |
Comparative Maps | |||||||
|---|---|---|---|---|---|---|---|---|
| Feature | Type | Position () | Map | Feature | Type | Position | Evidence | Actions |
| avgbs_15602 | locus | 9.1 | Oat - Oat-2018-Consensus - Oat-2018-Consensus_Mrg03 | avgbs_15602 | locus | 1.9 () | Automated name-based | View Maps |
| Oat - Oat-2016-Consensus - Oat-2016-Consensus-3 | avgbs_15602 | locus | 1.9 () | Automated name-based | View Maps | |||
| GMI_ES05_c23466_190 | locus | 6.8 | Oat - Oat-2017-U01BxU006 - Oat-2017-U01BxU006-29 | GMI_ES05_c23466_190 | locus | 20.17 (cM) | Automated name-based | View Maps |
| Oat - Oat-2018-Consensus - Oat-2018-Consensus_Mrg03 | GMI_ES05_c23466_190 | locus | 3.5 () | Automated name-based | View Maps | |||
| Oat - Oat-2014-CrownRust - Oat-2014-AsbxMN_30 | GMI_ES05_c23466_190 | locus | 0 (cM) | Automated name-based | View Maps | |||
| Oat - Oat-2016-DxE - Oat-2016-DxE_9b | GMI_ES05_c23466_190 | locus | 69.2 () | Automated name-based | View Maps | |||
| Oat - Oat-2016-AxM - Oat-2016-AxM_26 | GMI_ES05_c23466_190 | locus | 0 () | Automated name-based | View Maps | |||
| Oat - Oat-2016-OxT - Oat-2016-OxT_4b | GMI_ES05_c23466_190 | locus | 148.9 () | Automated name-based | View Maps | |||
| Oat - Oat-2016-BxG - Oat-2016-BxG_33 | GMI_ES05_c23466_190 | locus | 2.7 () | Automated name-based | View Maps | |||
| Oat - Oat-2016-IL4 - Oat-2016-IL4_19 | GMI_ES05_c23466_190 | locus | 9.8 () | Automated name-based | View Maps | |||
| Oat - Oat-2016-Consensus - Oat-2016-Consensus-3 | GMI_ES05_c23466_190 | locus | 3.5 () | Automated name-based | View Maps | |||
| Oat - Oat-2016-IL5 - Oat-2016-IL5_SS24 | GMI_ES05_c23466_190 | locus | 0.3 () | Automated name-based | View Maps | |||
| GMI_ES01_c14622_328 | locus | 6.8 | Oat - Oat-2016-IL5 - Oat-2016-IL5_SS24 | GMI_ES01_c14622_328 | locus | 0 () | Automated name-based | View Maps |
| Oat - Oat-2014-CrownRust - Oat-2014-AsbxMN_30 | GMI_ES01_c14622_328 | locus | 0 (cM) | Automated name-based | View Maps | |||
| Oat - Oat-2016-HxZ - Oat-2016-HxZ_ksh14 | GMI_ES01_c14622_328 | locus | 0 () | Automated name-based | View Maps | |||
| Oat - Oat-2016-BxG - Oat-2016-BxG_33 | GMI_ES01_c14622_328 | locus | 1.4 () | Automated name-based | View Maps | |||
| Oat - Oat-2016-OxT - Oat-2016-OxT_4b | GMI_ES01_c14622_328 | locus | 148.9 () | Automated name-based | View Maps | |||
| Oat - Oat-2017-U01BxU006 - Oat-2017-U01BxU006-29 | GMI_ES01_c14622_328 | locus | 22.33 (cM) | Automated name-based | View Maps | |||
| Oat - Oat-2016-IL4 - Oat-2016-IL4_19 | GMI_ES01_c14622_328 | locus | 9.8 () | Automated name-based | View Maps | |||
| Oat - Oat-2016-DxE - Oat-2016-DxE_9b | GMI_ES01_c14622_328 | locus | 69.2 () | Automated name-based | View Maps | |||
| Oat - Oat-2016-AxM - Oat-2016-AxM_26 | GMI_ES01_c14622_328 | locus | 0 () | Automated name-based | View Maps | |||
| Oat - Oat-2018-Consensus - Oat-2018-Consensus_Mrg03 | GMI_ES01_c14622_328 | locus | 3.3 () | Automated name-based | View Maps | |||
| Oat - Oat-2016-Consensus - Oat-2016-Consensus-3 | GMI_ES01_c14622_328 | locus | 3.3 () | Automated name-based | View Maps | |||
| avgbs_12200 | locus | 6.8 | Oat - Oat-2016-Consensus - Oat-2016-Consensus-3 | avgbs_12200 | locus | 3.4 () | Automated name-based | View Maps |
| Oat - Oat-2018-Consensus - Oat-2018-Consensus_Mrg03 | avgbs_12200 | locus | 3.4 () | Automated name-based | View Maps | |||
| avgbs_12201 | locus | 6.8 | Oat - Oat-2020-CORE-BG - Oat-2020-CORE-BG_Mrg03 | avgbs_12201 | locus | 0 (cM) | Automated name-based | View Maps |
| Oat - Oat-2016-Consensus - Oat-2016-Consensus-3 | avgbs_12201 | locus | 0.5 () | Automated name-based | View Maps | |||
| Oat - Oat-2018-Consensus - Oat-2018-Consensus_Mrg03 | avgbs_12201 | locus | 0.5 () | Automated name-based | View Maps | |||
| avgbs_16263 | locus | 2.3 | Oat - Oat-2016-Consensus - Oat-2016-Consensus-3 | avgbs_16263 | locus | 15.2 () | Automated name-based | View Maps |
| Oat - Oat-2018-Consensus - Oat-2018-Consensus_Mrg03 | avgbs_16263 | locus | 15.2 () | Automated name-based | View Maps | |||
| avgbs_121531 | locus | 2.3 | Oat - Oat-2018-Consensus - Oat-2018-Consensus_Mrg03 | avgbs_121531 | locus | 3.5 () | Automated name-based | View Maps |
| Oat - Oat-2016-Consensus - Oat-2016-Consensus-3 | avgbs_121531 | locus | 3.5 () | Automated name-based | View Maps | |||
| Oat - Oat-2016-OxT - Oat-2016-OxT_4b | avgbs_121531 | locus | 148.9 () | Automated name-based | View Maps | |||
| avgbs_120404 | locus | 2.3 | Oat - Oat-2018-Consensus - Oat-2018-Consensus_Mrg03 | avgbs_120404 | locus | 9 () | Automated name-based | View Maps |
| Oat - Oat-2016-Consensus - Oat-2016-Consensus-3 | avgbs_120404 | locus | 9 () | Automated name-based | View Maps | |||
| avgbs_121954 | locus | 2.3 | Oat - Oat-2018-Consensus - Oat-2018-Consensus_Mrg03 | avgbs_121954 | locus | 12.8 () | Automated name-based | View Maps |
| Oat - Oat-2016-Consensus - Oat-2016-Consensus-3 | avgbs_121954 | locus | 12.8 () | Automated name-based | View Maps | |||
| avgbs_207989 | locus | 2.3 | Oat - Oat-2016-Consensus - Oat-2016-Consensus-3 | avgbs_207989 | locus | 12.8 () | Automated name-based | View Maps |
| Oat - Oat-2018-Consensus - Oat-2018-Consensus_Mrg03 | avgbs_207989 | locus | 12.8 () | Automated name-based | View Maps | |||
| avgbs_80844 | locus | 2.3 | Oat - Oat-2016-OxT - Oat-2016-OxT_4b | avgbs_80844 | locus | 148.9 () | Automated name-based | View Maps |
| Oat - Oat-2018-Consensus - Oat-2018-Consensus_Mrg03 | avgbs_80844 | locus | 3.5 () | Automated name-based | View Maps | |||
| Oat - Oat-2016-Consensus - Oat-2016-Consensus-3 | avgbs_80844 | locus | 3.5 () | Automated name-based | View Maps | |||
| GMI_ES01_c15143_210 | locus | 2.3 | Oat - Oat-2016-DxE - Oat-2016-DxE_9b | GMI_ES01_c15143_210 | locus | 67.9 () | Automated name-based | View Maps |
| Oat - Oat-2016-OxT - Oat-2016-OxT_4b | GMI_ES01_c15143_210 | locus | 148.9 () | Automated name-based | View Maps | |||
| Oat - Oat-2016-PxG - Oat-2016-PxG_25 | GMI_ES01_c15143_210 | locus | 4.7 () | Automated name-based | View Maps | |||
| Oat - Oat-2017-UTauxU017 - Oat-2017-UTauxU017-11 | GMI_ES01_c15143_210 | locus | 0 (cM) | Automated name-based | View Maps | |||
| Oat - Oat-2018-Consensus - Oat-2018-Consensus_Mrg03 | GMI_ES01_c15143_210 | locus | 9 () | Automated name-based | View Maps | |||
| Oat - Oat-2016-HxZ - Oat-2016-HxZ_ksh14 | GMI_ES01_c15143_210 | locus | 0 () | Automated name-based | View Maps | |||
| Oat - Oat-2017-U01BxU006 - Oat-2017-U01BxU006-29 | GMI_ES01_c15143_210 | locus | 18.96 (cM) | Automated name-based | View Maps | |||
| Oat - Oat-2016-Consensus - Oat-2016-Consensus-3 | GMI_ES01_c15143_210 | locus | 9 () | Automated name-based | View Maps | |||
| Oat - Avena_2013_SNP - Avena_2013_SNP_10D-F-1 | GMI_ES01_c15143 | locus | 4.83 () | Automated name-based | View Maps | |||
| avgbs_202023 | locus | 2.3 | Oat - Oat-2016-Consensus - Oat-2016-Consensus-3 | avgbs_202023 | locus | 15.2 () | Automated name-based | View Maps |
| Oat - Oat-2018-Consensus - Oat-2018-Consensus_Mrg03 | avgbs_202023 | locus | 15.2 () | Automated name-based | View Maps | |||
| avgbs_107747 | locus | 2.3 | Oat - Oat-2016-OxT - Oat-2016-OxT_4b | avgbs_107747 | locus | 143.5 () | Automated name-based | View Maps |
| Oat - Oat-2016-Consensus - Oat-2016-Consensus-3 | avgbs_107747 | locus | 3.3 () | Automated name-based | View Maps | |||
| Oat - Oat-2018-Consensus - Oat-2018-Consensus_Mrg03 | avgbs_107747 | locus | 3.3 () | Automated name-based | View Maps | |||
| ISU1254A | locus | 0 | Oat - Oat-2016-Consensus - Oat-2016-Consensus-3 | ISU1254A | locus | 6.1 () | Automated name-based | View Maps |
| Oat - Oat-2018-Consensus - Oat-2018-Consensus_Mrg03 | ISU1254A | locus | 6.1 () | Automated name-based | View Maps | |||
| Oat - Oat-2009-KxO - Oat-2009-KxO_19_25_27 | ISU1254A | locus | 3 (cM) | Automated name-based | View Maps | |||
| Oat - Oat, KxO, 2003 - Oat-KxO/2003-19+27 | isu1254a | locus | 0 () | Automated name-based | View Maps | |||
| Oat - Oat-2013-KxO-RGA - RGA-2013_KO19_27 | isu1254a | locus | 0 (cM) | Automated name-based | View Maps | |||
| CDO420B | locus | 0 | Oat - Oat-2016-Consensus - Oat-2016-Consensus-3 | CDO420B | locus | 5.1 () | Automated name-based | View Maps |
| Barley - Barley consensus 2 - Barley-Consensus2-7H | CDO420B | locus | 127.7 () | Automated name-based, Automated name-based | View Maps | |||
| Oat - Oat, KxO, 2003 - Oat-KxO/2003-19+27 | CDO420B | locus | 0.78 () | Automated name-based | View Maps | |||
| Oat - Oat-2018-Consensus - Oat-2018-Consensus_Mrg03 | CDO420B | locus | 5.1 () | Automated name-based | View Maps | |||
| Oat - Oat-2009-KxO - Oat-2009-KxO_19_25_27 | CDO420B | locus | 6 (cM) | Automated name-based | View Maps | |||
| Barley - Barley, BinMap 2005 - Hordeum-Bins-7H | CDO420B | locus | 134 () | Automated name-based | View Maps | |||
| Oat - Oat-2013-KxO-RGA - RGA-2013_KO19_27 | cdo420b | locus | 1 (cM) | Automated name-based | View Maps | |||
| ISU1247A | locus | 0 | Oat - Oat-2009-KxO - Oat-2009-KxO_19_25_27 | ISU1247A | locus | 3 (cM) | Automated name-based | View Maps |
| Oat - Oat-2016-Consensus - Oat-2016-Consensus-3 | ISU1247A | locus | 6.1 () | Automated name-based | View Maps | |||
| Oat - Oat, KxO, 2003 - Oat-KxO/2003-19+27 | ISU1247A | locus | 0 () | Automated name-based | View Maps | |||
| Oat - Oat-2018-Consensus - Oat-2018-Consensus_Mrg03 | ISU1247A | locus | 6.1 () | Automated name-based | View Maps | |||
| Diploid oat - Diploid Oat, Iowa St. - Avena-Wise-D | ISU1247A | locus | 446.8 () | Automated name-based | View Maps | |||

GrainGenes is a product of the US Department of Agriculture.
| | ||
