|
|
[ Download Map Data ]
[ Download Feature Correspondence Data ]
|
|
|
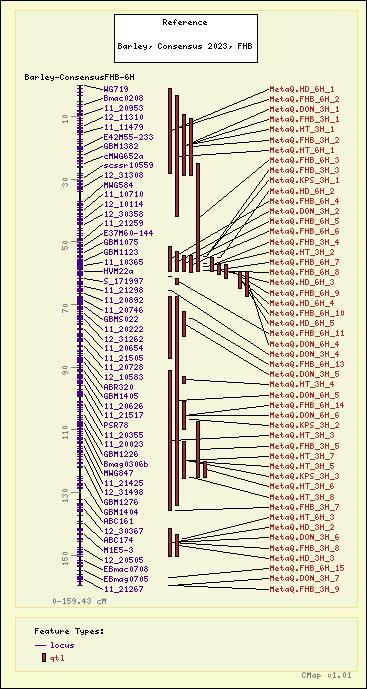
Barley - Barley, Consensus 2023, FHB - Barley-ConsensusFHB-6H
(Click headers to resort) |
Comparative Maps |
| Feature |
Type |
Position (cM) |
Map |
Feature |
Type |
Position |
Evidence |
Actions |
|
11_20858
|
locus
|
0
|
Barley - Barley, 2012 9KSNP v2 - Barley2012_9KSNPv2-3H
|
11_20858
|
locus
|
0 (cM)
|
Automated name-based
|
View Maps
|
|
12_31428
|
locus
|
0
|
Barley - Barley, 2012 9KSNP v2 - Barley2012_9KSNPv2-3H
|
12_31428
|
locus
|
0 (cM)
|
Automated name-based
|
View Maps
|
|
M1E4-11
|
locus
|
0.19
|
Barley - Barley, Zhedar2, FHB QTL - Hordeum-Zhedar2-FHB-QTL-3H
|
M1E4-11
|
locus
|
0 ()
|
Automated name-based
|
View Maps
|
|
M5E3-13
|
locus
|
8.19
|
Barley - Barley, Zhedar2, FHB QTL - Hordeum-Zhedar2-FHB-QTL-3H
|
M5E3-13
|
locus
|
8.3 ()
|
Automated name-based
|
View Maps
|
|
12_10103
|
locus
|
8.39
|
Barley - Barley, 2012 9KSNP v2 - Barley2012_9KSNPv2-3H
|
12_10103
|
locus
|
1.6 (cM)
|
Automated name-based
|
View Maps
|
|
12_20090
|
locus
|
8.39
|
No other positions |
|
11_20797
|
locus
|
8.39
|
Barley - Barley, 2012 9KSNP v2 - Barley2012_9KSNPv2-3H
|
11_20797
|
locus
|
1.6 (cM)
|
Automated name-based
|
View Maps
|
|
11_20159
|
locus
|
8.39
|
No other positions |
|
12_11434
|
locus
|
8.39
|
No other positions |
|
12_31448
|
locus
|
8.39
|
No other positions |
|
11_11453
|
locus
|
8.39
|
Barley - Barley, 2012 9KSNP v2 - Barley2012_9KSNPv2-3H
|
11_11453
|
locus
|
2.4 (cM)
|
Automated name-based
|
View Maps
|
|
11_20252
|
locus
|
8.39
|
Barley - Barley, 2012 9KSNP v2 - Barley2012_9KSNPv2-3H
|
11_20252
|
locus
|
2.7 (cM)
|
Automated name-based
|
View Maps
|
|
12_10173
|
locus
|
8.39
|
Barley - Barley, 2012 9KSNP v2 - Barley2012_9KSNPv2-3H
|
12_10173
|
locus
|
2.7 (cM)
|
Automated name-based
|
View Maps
|
|
ABG070
|
locus
|
8.39
|
Barley - Barley, Steptoe x Morex, SNP - Hordeum-StMo-SNP-3H
|
ABG070
|
locus
|
0 ()
|
Automated name-based
|
View Maps
|
|
Barley - Barley, Consensus 2005, SNP - Hordeum-Consensus2005-SNP-3H
|
ABG070
|
locus
|
0 ()
|
Automated name-based
|
View Maps
|
|
Barley - Barley, Agronomic QTL Consensus - Hordeum-QTLConsensus-Agronomic-3
|
ABG070
|
locus
|
0 ()
|
Automated name-based
|
View Maps
|
|
Barley - Barley, Abiotic QTL Consensus - Hordeum-QTLConsensus-Abiotic-3H
|
ABG070
|
locus
|
0 ()
|
Automated name-based
|
View Maps
|
|
Barley - Barley, Pilot OPA1, Consensus - Hordeum-PilotOPA1-3H
|
ABG070
|
locus
|
0 ()
|
Automated name-based
|
View Maps
|
|
Barley - Barley, AxK - Hordeum-AxK-3H
|
ABG070
|
locus
|
0 ()
|
Automated name-based
|
View Maps
|
|
Barley - Barley, OPA 2009, Consensus - Hordeum-OPA-2009-3H
|
ABG070
|
locus
|
0 ()
|
Automated name-based
|
View Maps
|
|
Barley - Barley, Integrated, Marcel 2009 - Hv-Integrated2009-3H
|
ABG070
|
locus
|
12.7 ()
|
Automated name-based
|
View Maps
|
|
Barley - Barley, BinMap 2005 - Hordeum-Bins-3H
|
ABG070
|
locus
|
0.8 ()
|
Automated name-based
|
View Maps
|
|
Barley - Barley, Consensus 2007, SSR - Hordeum-Consensus2007-SSR-3H
|
ABG070
|
locus
|
4.66 ()
|
Automated name-based
|
View Maps
|
|
GBM1280
|
locus
|
8.39
|
Barley - Barley, Integrated, Marcel 2009 - Hv-Integrated2009-3H
|
GBM1280
|
locus
|
9.6 ()
|
Automated name-based
|
View Maps
|
|
Barley - Barley, Consensus 2007, SSR - Hordeum-Consensus2007-SSR-3H
|
GBM1280
|
locus
|
3.83 ()
|
Automated name-based
|
View Maps
|
|
Barley - Barley, Consensus 2006, Stein - Hv-Consensus2006-Stein-3H
|
GBM1280-3H
|
locus
|
9.2 ()
|
Automated name-based
|
View Maps
|
|
11_20953
|
locus
|
9.39
|
No other positions |
|
12_31409
|
locus
|
9.39
|
Barley - Barley, 2012 9KSNP v2 - Barley2012_9KSNPv2-3H
|
12_31409
|
locus
|
3.1 (cM)
|
Automated name-based
|
View Maps
|
|
ABG316a
|
locus
|
9.39
|
Barley - Barley, Consensus 2006, Marcel - Hv-Consensus2006-Marcel-3H
|
ABG316A
|
locus
|
12.72 ()
|
Automated name-based
|
View Maps
|
|
Barley - Barley, Agronomic QTL Consensus - Hordeum-QTLConsensus-Agronomic-3
|
ABG316A
|
locus
|
3.22 ()
|
Automated name-based
|
View Maps
|
|
Barley - Barley, Consensus 2005, SNP - Hordeum-Consensus2005-SNP-3H
|
ABG316A
|
locus
|
3.22 ()
|
Automated name-based
|
View Maps
|
|
Barley - Barley, SxM, Feed QTLs - Hordeum-SxM-FeedQTL-3H
|
ABG316A
|
locus
|
2.1 ()
|
Automated name-based
|
View Maps
|
|
Barley - Barley, Abiotic QTL Consensus - Hordeum-QTLConsensus-Abiotic-3H
|
ABG316A
|
locus
|
3.22 ()
|
Automated name-based
|
View Maps
|
|
Barley - Barley consensus 2 - Barley-Consensus2-3H
|
ABG316A
|
locus
|
1.4 ()
|
Automated name-based
|
View Maps
|
|
Barley - Barley, SxM - Hordeum-NABGMP1-3H
|
ABG316A
|
locus
|
2.1 ()
|
Automated name-based
|
View Maps
|
|
Barley - Barley, BinMap 2005 - Hordeum-Bins-3H
|
ABG316A
|
locus
|
4.9 ()
|
Automated name-based
|
View Maps
|
|
Barley - Barley, Steptoe x Morex, SNP - Hordeum-StMo-SNP-3H
|
ABG316A
|
locus
|
3.37 ()
|
Automated name-based
|
View Maps
|
|
Barley - Barley, Steptoe x Morex, SSR - Hordeum-SxM-SSR-3H
|
ABG316A
|
locus
|
0 ()
|
Automated name-based
|
View Maps
|
|
Barley - Barley, Integrated, Marcel 2009 - Hv-Integrated2009-3H
|
ABG316A
|
locus
|
16.1 ()
|
Automated name-based
|
View Maps
|
|
ABC171
|
locus
|
10.58
|
Barley - Barley, SxM basemap - Hordeum-SxM-base-3H
|
ABC171
|
locus
|
16.6 ()
|
Automated name-based
|
View Maps
|
|
Barley - Barley consensus 2003 - Barley-consensus-2003-2H
|
ABC171A
|
locus
|
144.8 ()
|
Automated name-based
|
View Maps
|
|
Barley - Barley consensus 2 - Barley-Consensus2-3H
|
ABC171
|
locus
|
9.6 ()
|
Automated name-based
|
View Maps
|
|
Barley - Barley, HxT - Hordeum-NABGMP2-3H
|
ABC171
|
locus
|
0 ()
|
Automated name-based
|
View Maps
|
|
Barley - Barley consensus - Barley-Consensus-3H
|
ABC171
|
locus
|
-29.4 ()
|
Automated name-based
|
View Maps
|
|
Barley - Barley, Steptoe x Morex, SSAP - Hordeum-SxM-SSAP-3H
|
ABC171
|
locus
|
16.9 ()
|
Automated name-based
|
View Maps
|
|
Barley - Barley, CxH - Hordeum-CxH-3H
|
ABC171
|
locus
|
14.08 ()
|
Automated name-based
|
View Maps
|
|
Barley - Barley, Consensus 2006, DArT - Hordeum-Consensus2006-DArT-3H
|
ABC171
|
locus
|
22.5 ()
|
Automated name-based
|
View Maps
|
|
Barley - Barley, SxM - Hordeum-NABGMP1-3H
|
ABC171
|
locus
|
16.3 ()
|
Automated name-based
|
View Maps
|
|
Barley - Barley, Zhedar2, FHB QTL - Hordeum-Zhedar2-FHB-QTL-3H
|
ABC171
|
locus
|
24.8 ()
|
Automated name-based
|
View Maps
|
|
Barley - Barley, HxT basemap - Hordeum-HxT-base-3H
|
ABC171
|
locus
|
0 ()
|
Automated name-based
|
View Maps
|
|
Barley - Barley, SxM, Feed QTLs - Hordeum-SxM-FeedQTL-3H
|
ABC171
|
locus
|
16.3 ()
|
Automated name-based
|
View Maps
|
|
11_20529
|
locus
|
10.82
|
Barley - Barley, 2012 9KSNP v2 - Barley2012_9KSNPv2-3H
|
11_20529
|
locus
|
3.3 (cM)
|
Automated name-based
|
View Maps
|
|
11_21190
|
locus
|
10.82
|
No other positions |
|
11_21398
|
locus
|
10.82
|
Barley - Barley, 2012 9KSNP v2 - Barley2012_9KSNPv2-3H
|
11_21398
|
locus
|
5.3 (cM)
|
Automated name-based
|
View Maps
|
|
12_11005
|
locus
|
10.82
|
No other positions |
|
12_30297
|
locus
|
10.82
|
No other positions |
|
11_21027
|
locus
|
12.18
|
No other positions |