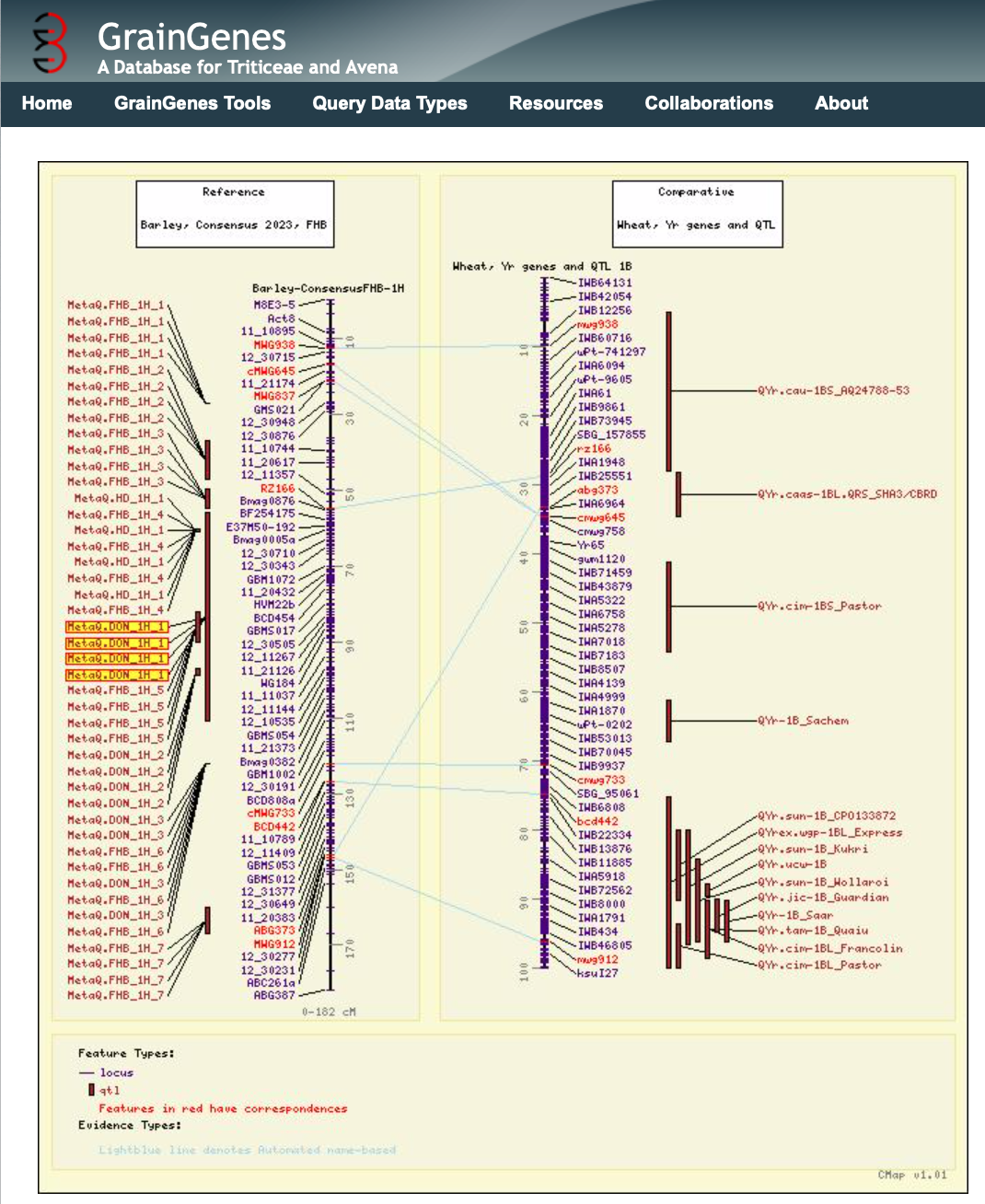
GrainGenes Reference : PBR-143-2 MapData : Barley, Consensus 2023, FHB
256 MetaQTL for FHB resistance, DON accumulation, and agronomic traits from 12 mapping studies were reported by Sallam, Haas, Huang, Tandukar, Muehlbauer, Smith, & Steffenson (2023) Meta-analysis of the genetics of resistance to Fusarium head blight and deoxynivalenol accumulation in barley and considerations for breeding. Plant Breeding 143:2. https://doi.org/10.1111/pbr.13121.
QTL were curated into the GrainGenes database and genetic maps were created for the CMap comparative mapping tool at GrainGenes. Users can now view a barley map for Fusarium traits and compare to, for example, the homologous wheat chromosome with stripe rust data, as shown below. CMap has been rigorously used prior to the current era of genome browsers, and can lend a keen insight into genetically mapped chromosome regions that share similar traits across the Triticeae. There are many tutorials for CMap in the GrainGenes Collection and new users are encouraged to explore that powerful genetic mapping tool.