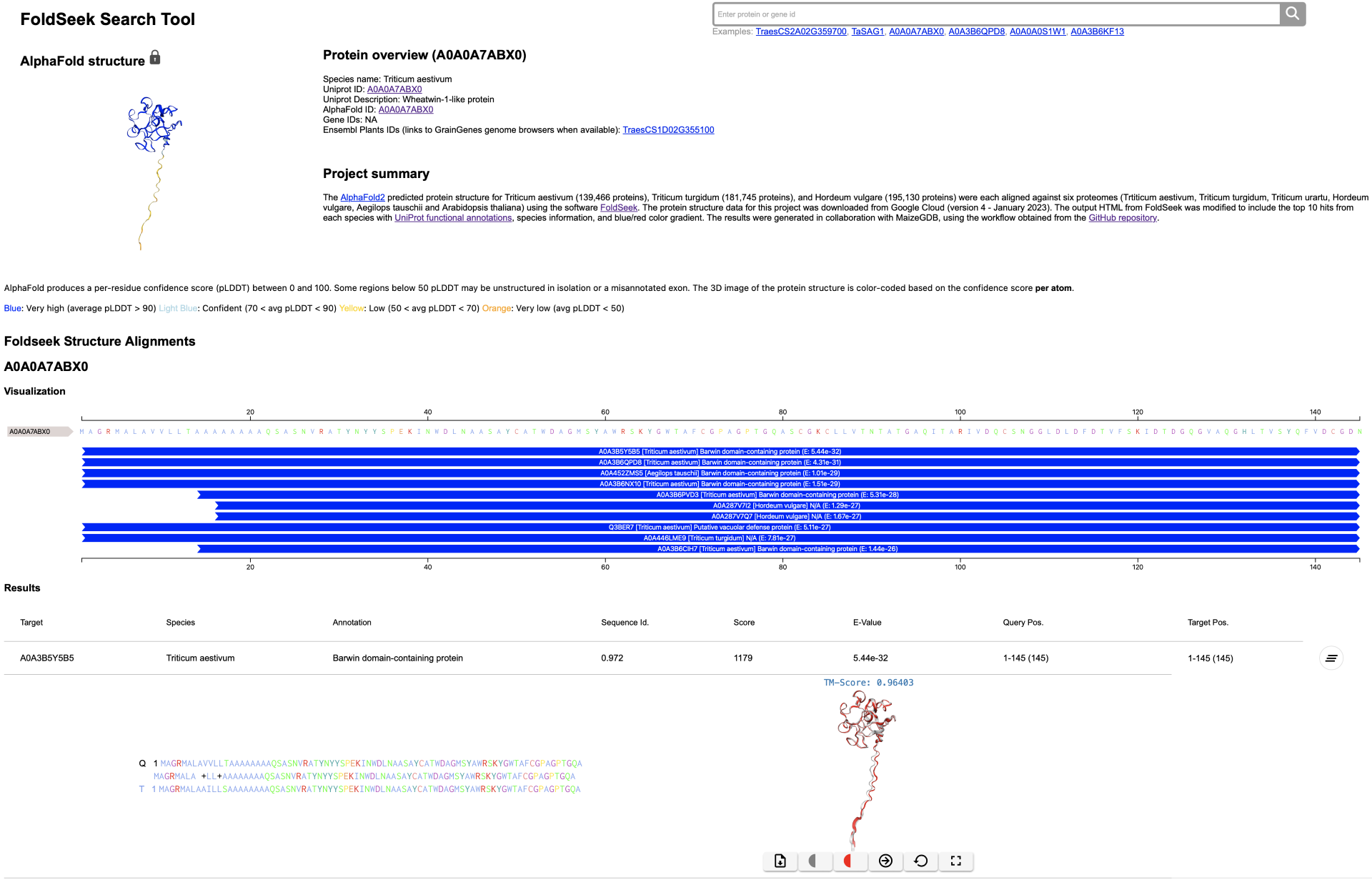
We added the FoldSeek Search Tool for AlphaFold2 structures of three pre-calculated genomes searched against six genomes. The predicted AlphaFold2 database structures for Triticum aestivum (139,466 proteins), Triticum turgidum (181,745 proteins), and Hordeum vulgare (195,130 proteins) were each aligned against six proteomes (T. aestivum, T. turgidum, Triticum urartu, H. vulgare, Aegilops tauschii and Arabidopsis thaliana) using the software FoldSeek. The protein structure data for this project was downloaded from Google Cloud (version 4 - January 2023). The output HTML from FoldSeek was modified to include the top 10 hits from each species with UniProt functional annotations, species information, and blue/red color gradient. The results were generated in collaboration with MaizeGDB, using the workflow obtained from the CrossFoldDB GitHub repository.
Users can search through the FoldSeek results using AlphaFold2/UniProt IDs or available EnsemblPlants IDs:
Result